Chapter 11 Visualization
XMAS 2.0 provides mulitple functions for visualization. For instance, using plot_volcano to display the results of differential analysis.
Outline of this Chapter:
11.2 Importing Data
data("amplicon_ps")
amplicon_ps_rarefy <- norm_rarefy(object = amplicon_ps,
size = 1114)
amplicon_ps_rarefy## phyloseq-class experiment-level object
## otu_table() OTU Table: [ 2308 taxa and 34 samples ]
## sample_data() Sample Data: [ 34 samples by 8 sample variables ]
## tax_table() Taxonomy Table: [ 2308 taxa by 7 taxonomic ranks ]
## phy_tree() Phylogenetic Tree: [ 2308 tips and 2306 internal nodes ]11.3 plot_boxplot
- calculate alpha diversity
dat_alpha <- run_alpha_diversity(ps = amplicon_ps_rarefy,
measures = c("Shannon", "Chao1", "Observed"))
head(dat_alpha)## TempRowNames SampleType Year Month Day Subject ReportedAntibioticUsage DaysSinceExperimentStart Description Observed Chao1
## 1 L1S140 gut 2008 10 28 2 Yes 0 2_Fece_10_28_2008 27 74.50
## 2 L1S208 gut 2009 1 20 2 No 84 2_Fece_1_20_2009 40 148.75
## 3 L1S8 gut 2008 10 28 1 Yes 0 1_Fece_10_28_2008 19 54.00
## 4 L1S281 gut 2009 4 14 2 No 168 2_Fece_4_14_2009 60 256.00
## 5 L3S242 right palm 2008 10 28 1 Yes 0 1_R_Palm_10_28_2008 16 42.00
## 6 L2S309 left palm 2009 1 20 2 No 84 2_L_Palm_1_20_2009 12 57.00
## se.chao1 Shannon
## 1 30.70970 3.126005
## 2 62.86107 3.303186
## 3 25.57190 2.688337
## 4 93.42352 3.664947
## 5 19.97805 2.692311
## 6 30.06061 2.082828plot_boxplot has many parameters, and help you enjoy it.
- single measure
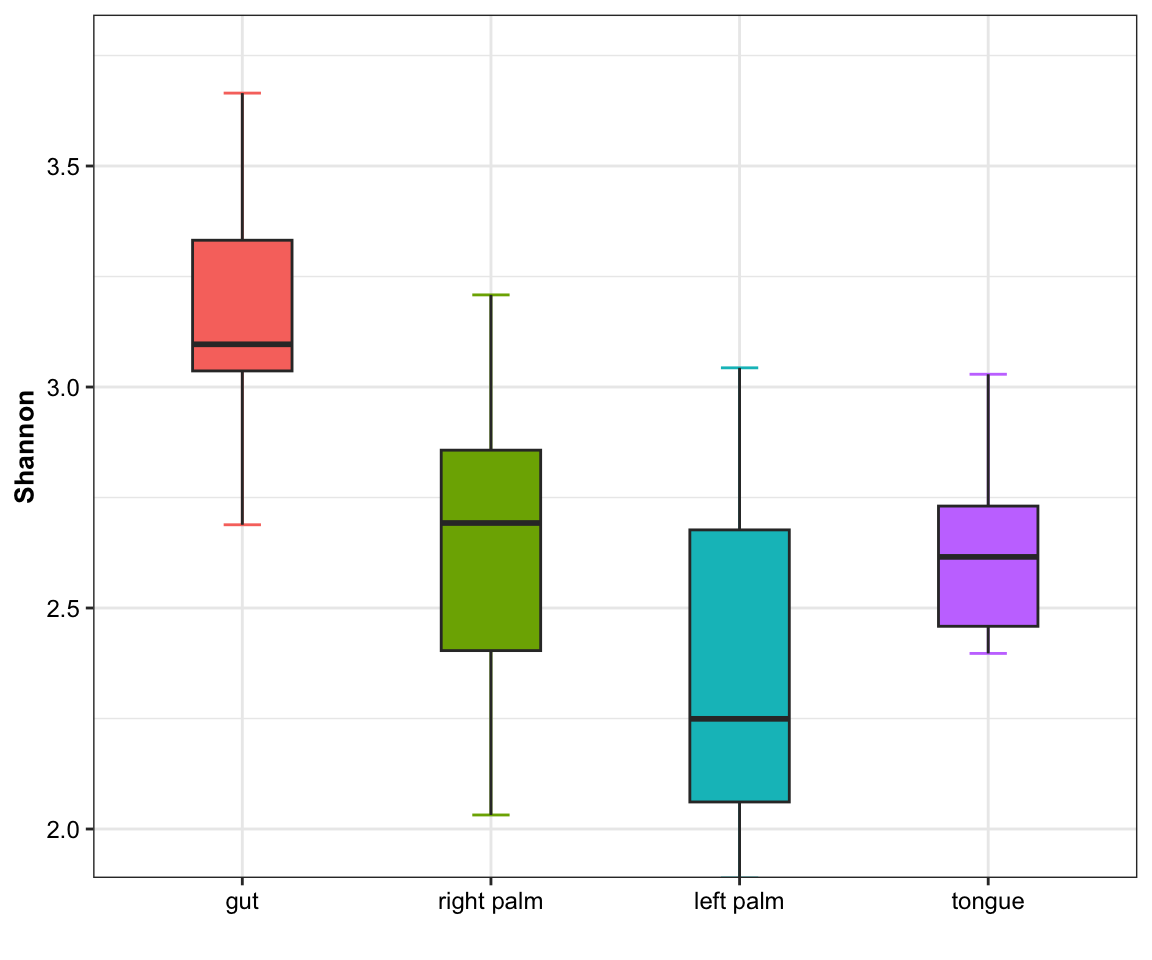
Figure 11.1: boxplot(single measure)
- single measure with significant results
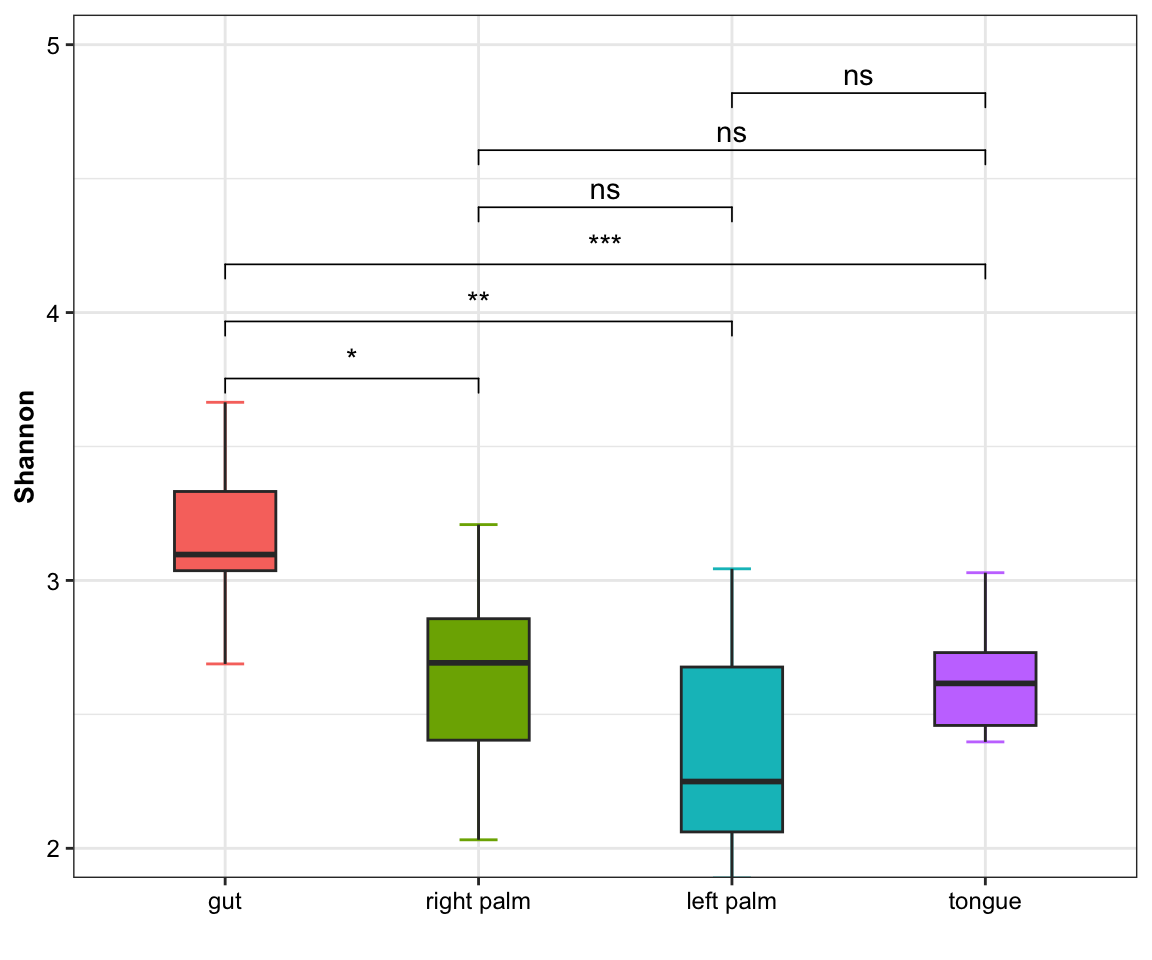
Figure 11.2: boxplot(single measure with significant results)
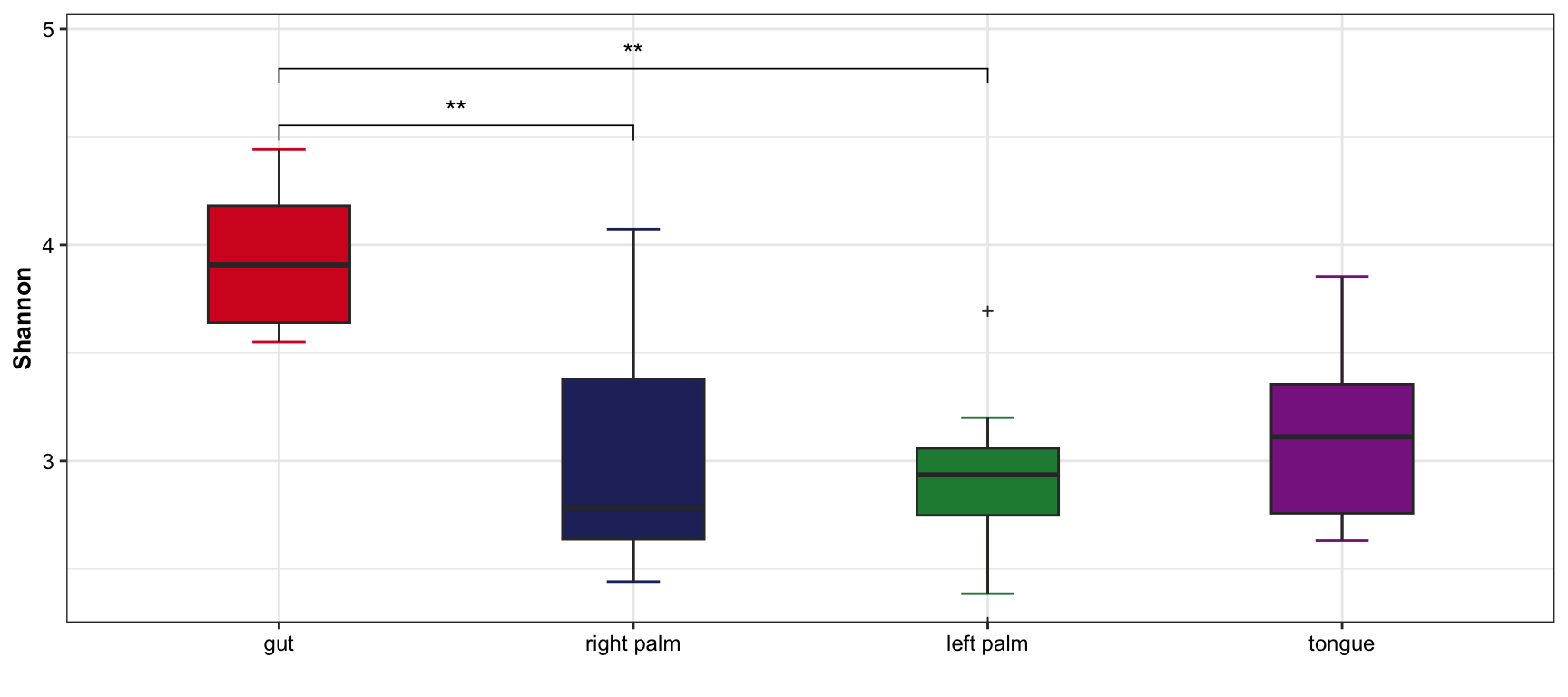
- single measure with significant results of pairwises
plot_boxplot(data = dat_alpha,
y_index = "Shannon",
group = "SampleType",
do_test = TRUE,
cmp_list = list(c("gut", "right palm"),
c("gut", "tongue")))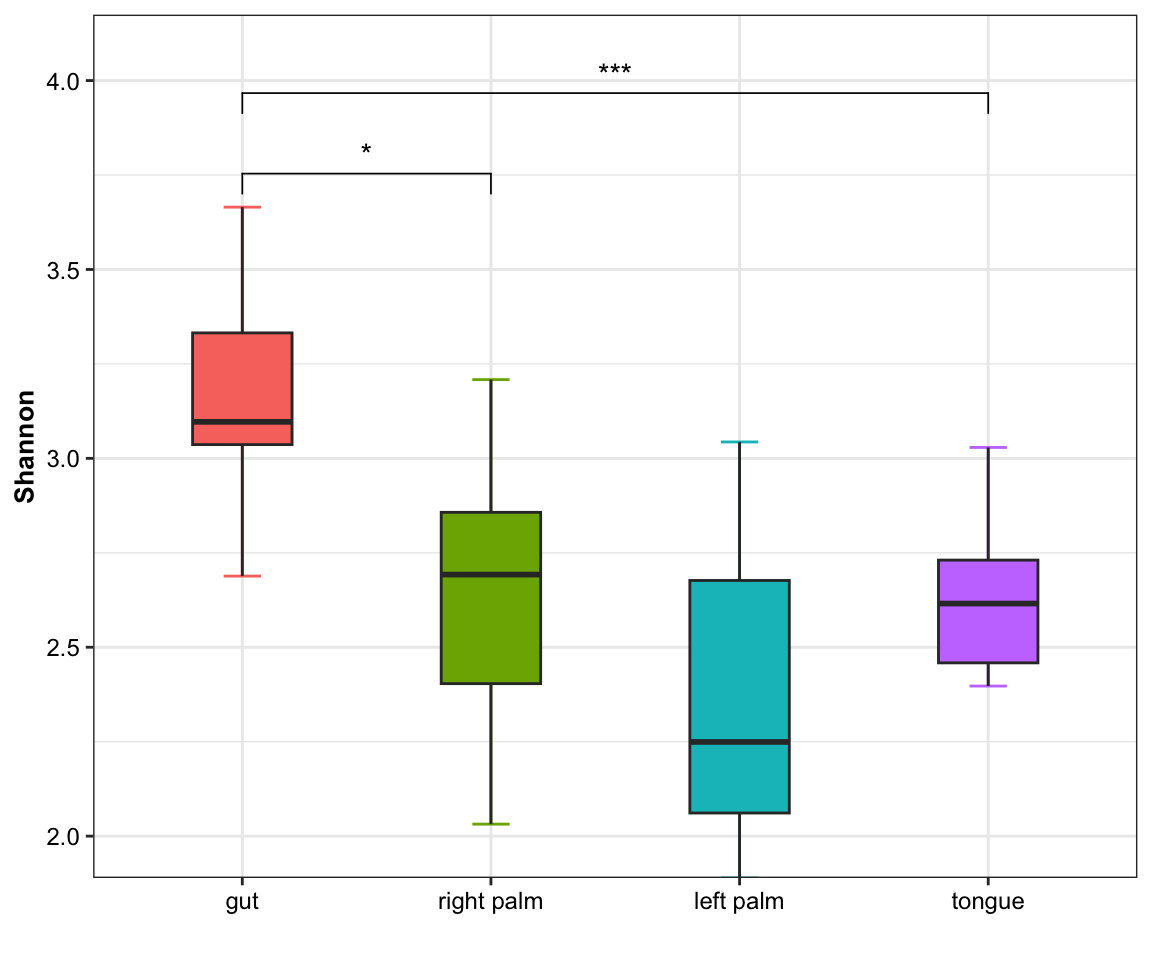
Figure 11.3: boxplot(single measure with significant results of pairwises)
- single measure with significant results of pairwises and outlier
plot_boxplot(data = dat_alpha,
y_index = "Shannon",
group = "SampleType",
do_test = TRUE,
cmp_list = list(c("gut", "right palm"),
c("gut", "tongue")),
outlier = TRUE)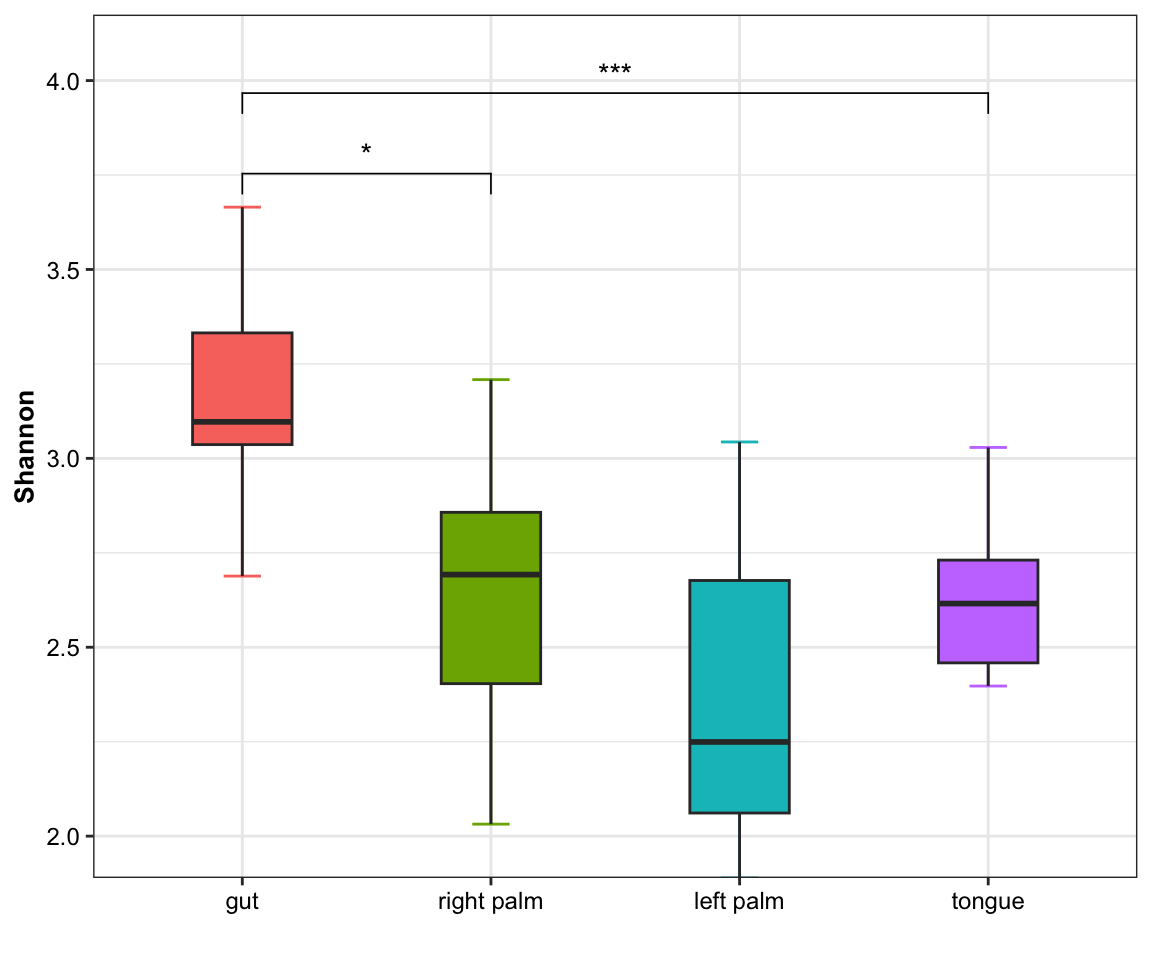
Figure 11.4: boxplot(single measure with significant results of pairwises and outlier)
- single measure with significant results of ref_group
plot_boxplot(data = dat_alpha,
y_index = "Shannon",
group = "SampleType",
do_test = TRUE,
ref_group = "gut")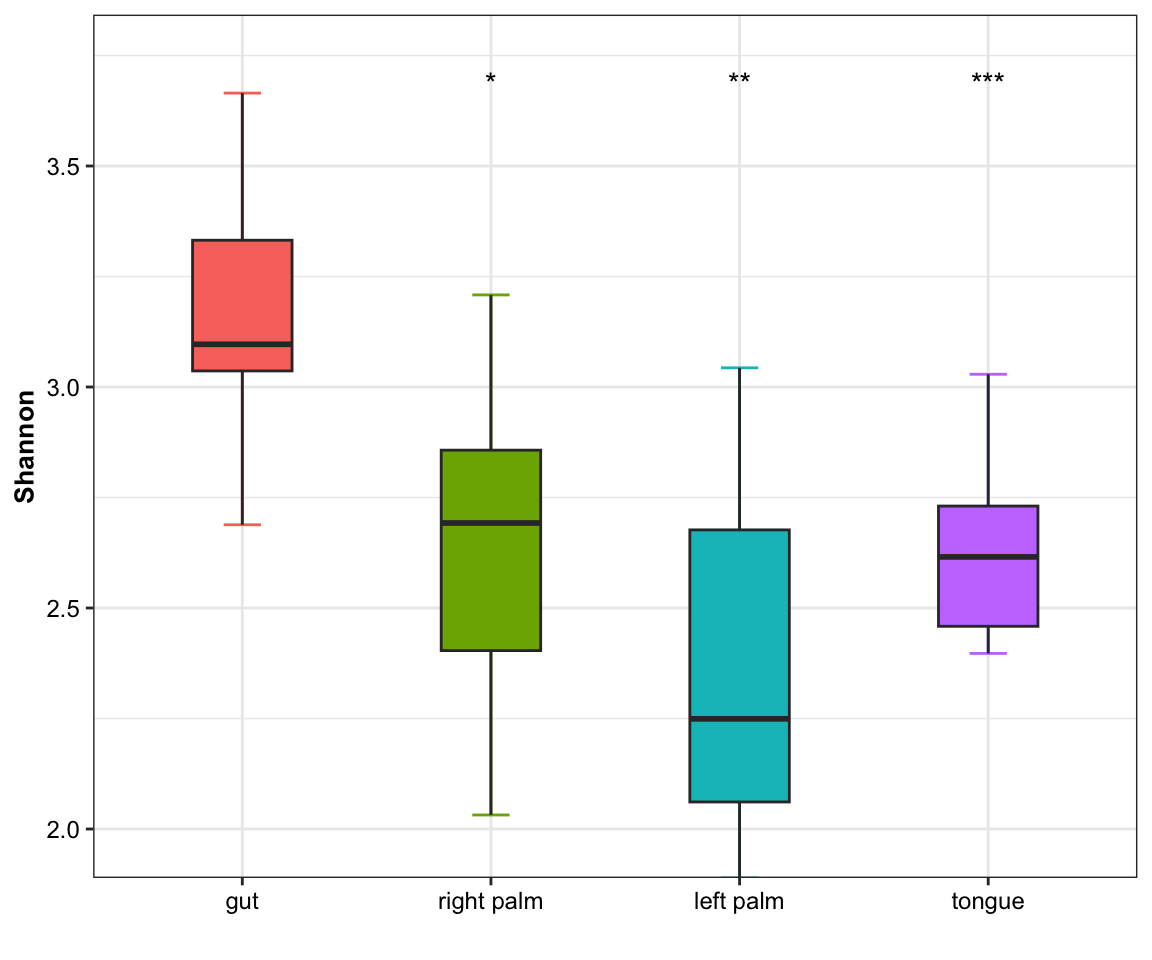
Figure 11.5: boxplot(single measure with significant results of ref_group)
- multiple measures
plot_boxplot(data = dat_alpha,
y_index = c("Shannon", "Chao1", "Observed"),
group = "SampleType",
group_names = c("gut", "right palm", "tongue"),
group_color = c("red", "green", "blue"),
ref_group = "gut",
method = "wilcox.test",
outlier = TRUE)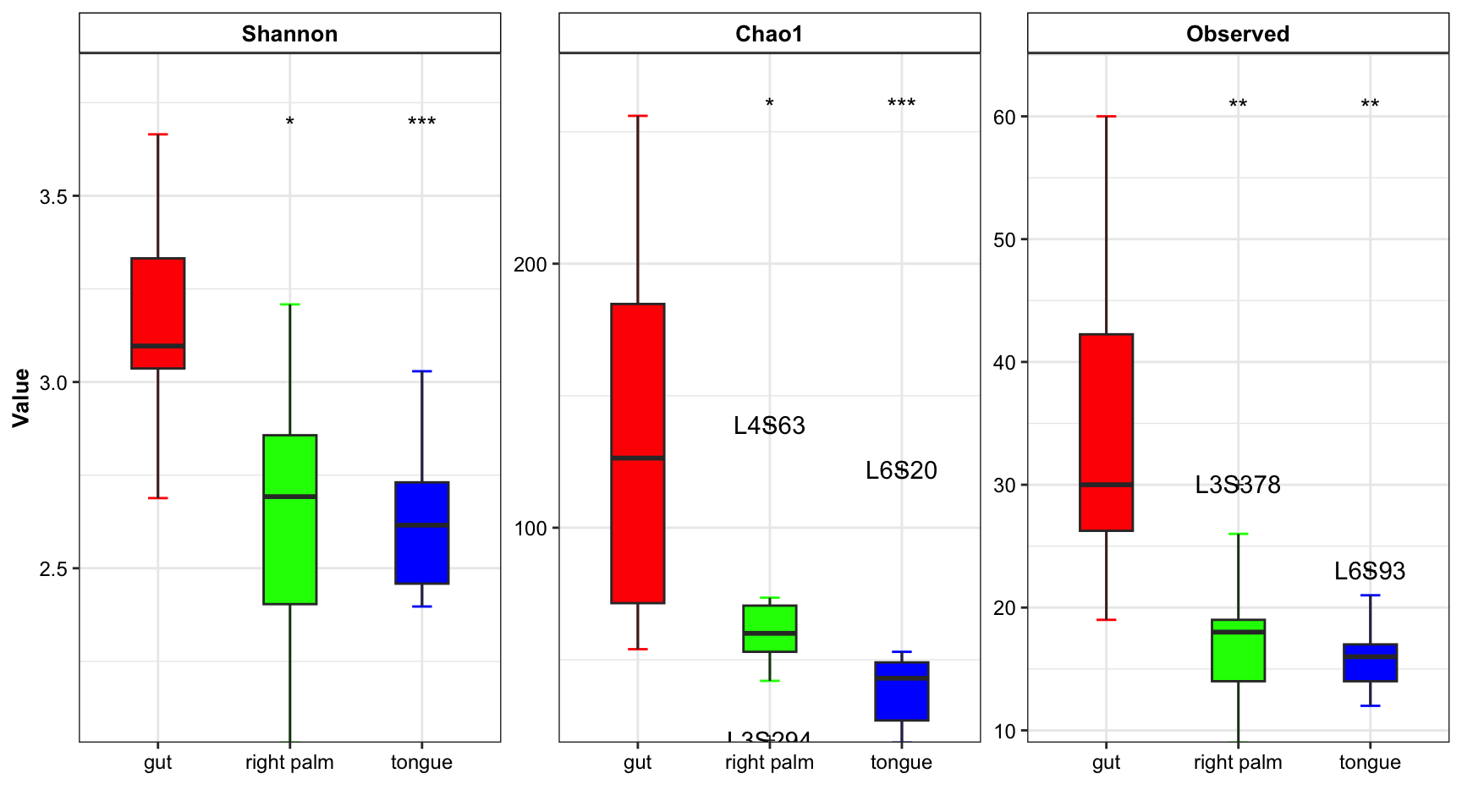
Figure 11.6: boxplot(multiple measure with group number)
- show group_number in the x-axis break
plot_boxplot(data = dat_alpha,
y_index = c("Shannon", "Chao1", "Observed"),
group = "SampleType",
group_names = c("gut", "right palm", "tongue"),
group_color = c("red", "green", "blue"),
do_test = TRUE,
method = "wilcox.test",
group_number = TRUE)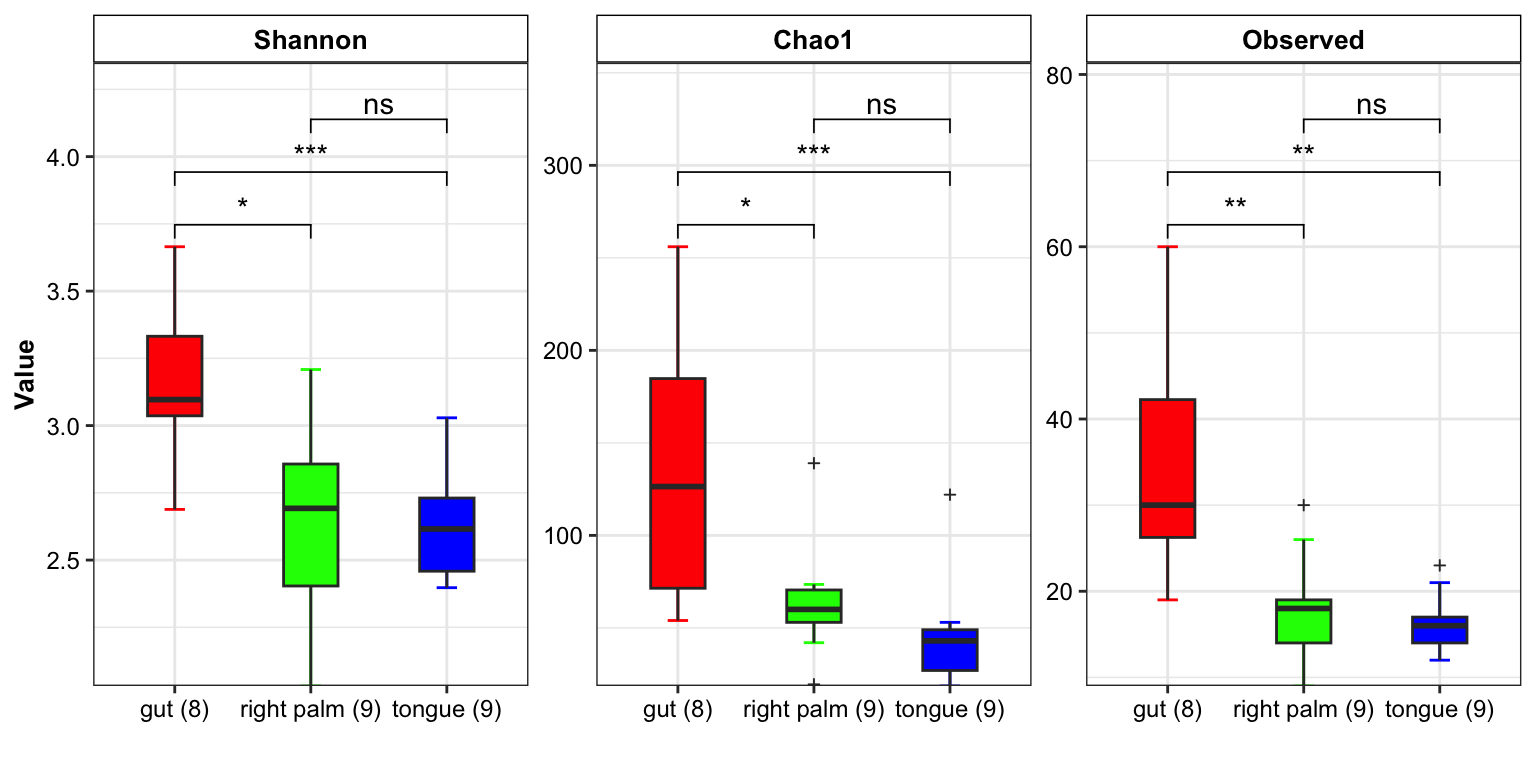
Figure 11.7: boxplot(group_number)
11.4 plot_barplot
plot_barplot has many parameters, and help you enjoy it.
- single measure
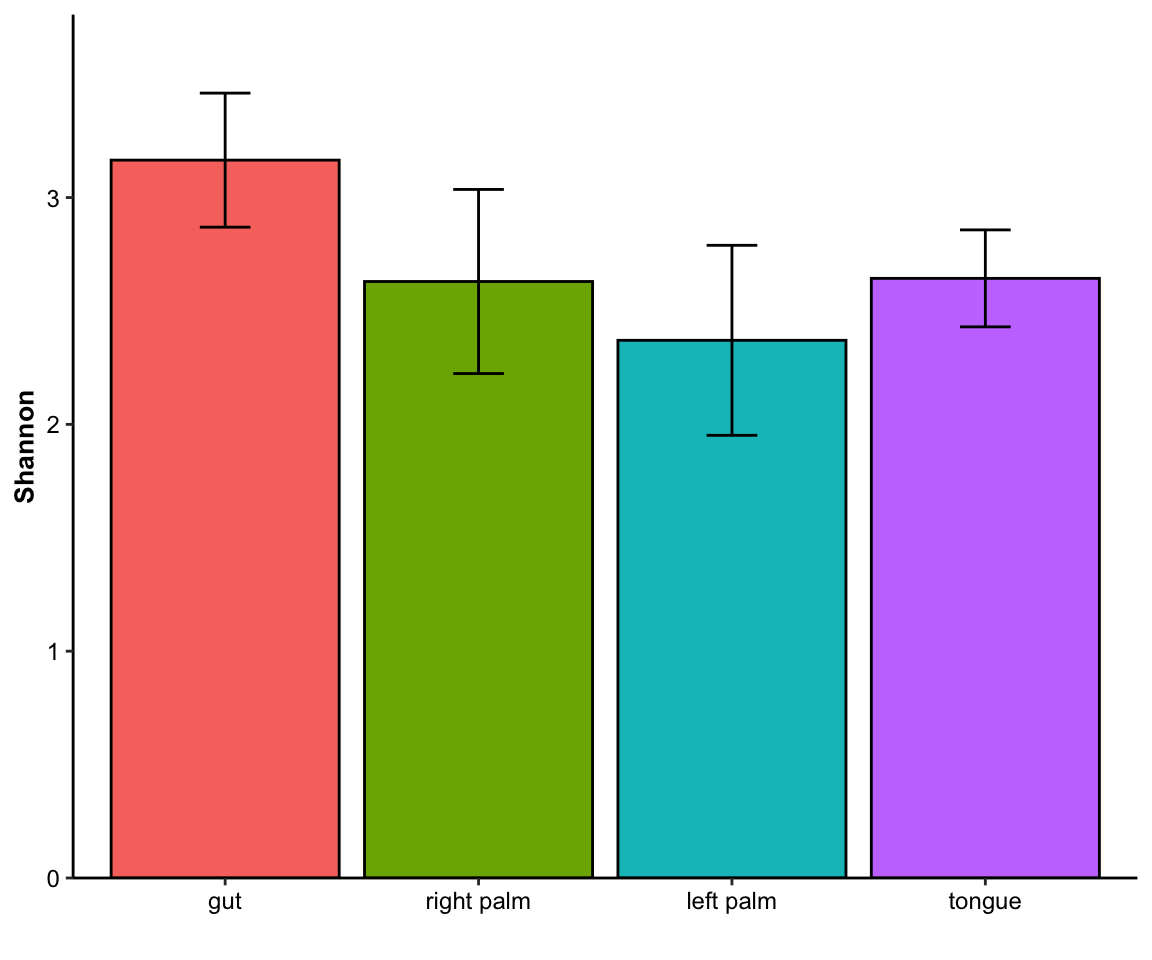
Figure 11.8: barplot(single measure)
- single measure with significant results
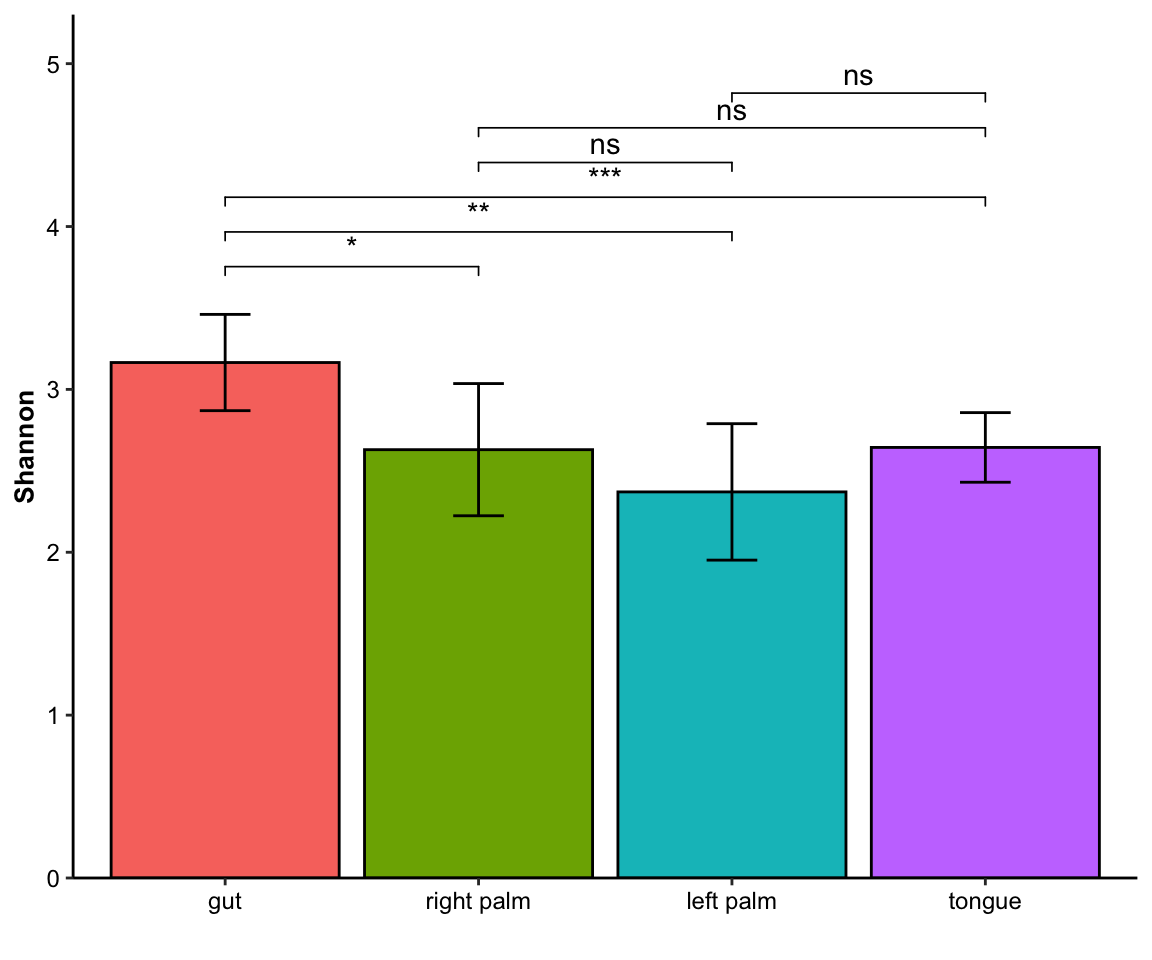
Figure 11.9: barplot(single measure with significant results)
- single measure with significant results of pairwises
plot_barplot(data = dat_alpha,
y_index = "Shannon",
group = "SampleType",
do_test = TRUE,
cmp_list = list(c("gut", "right palm"), c("gut", "tongue")))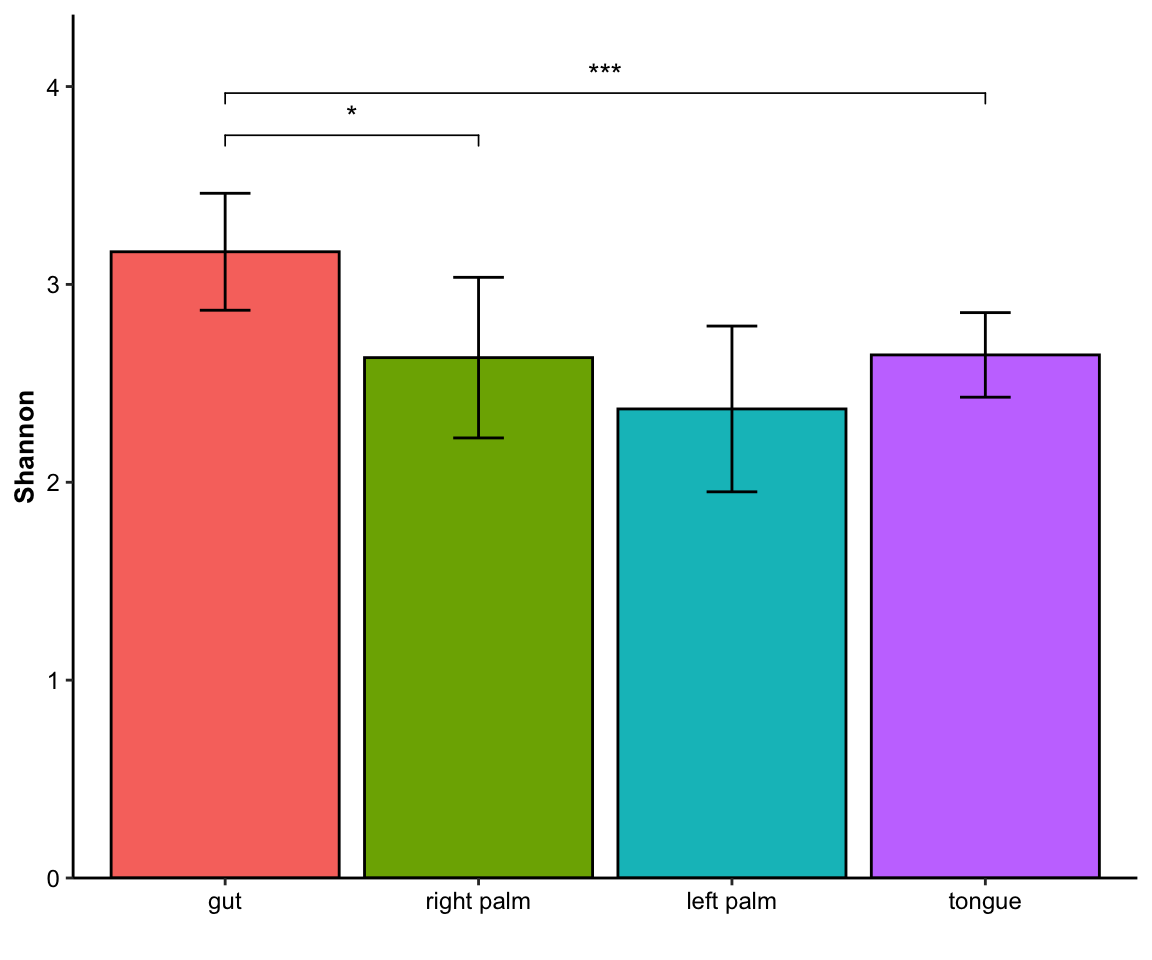
Figure 11.10: barplot(single measure with significant results of pairwises)
- single measure with significant results of ref_group
plot_barplot(data = dat_alpha,
y_index = "Shannon",
group = "SampleType",
do_test = TRUE,
ref_group = "gut")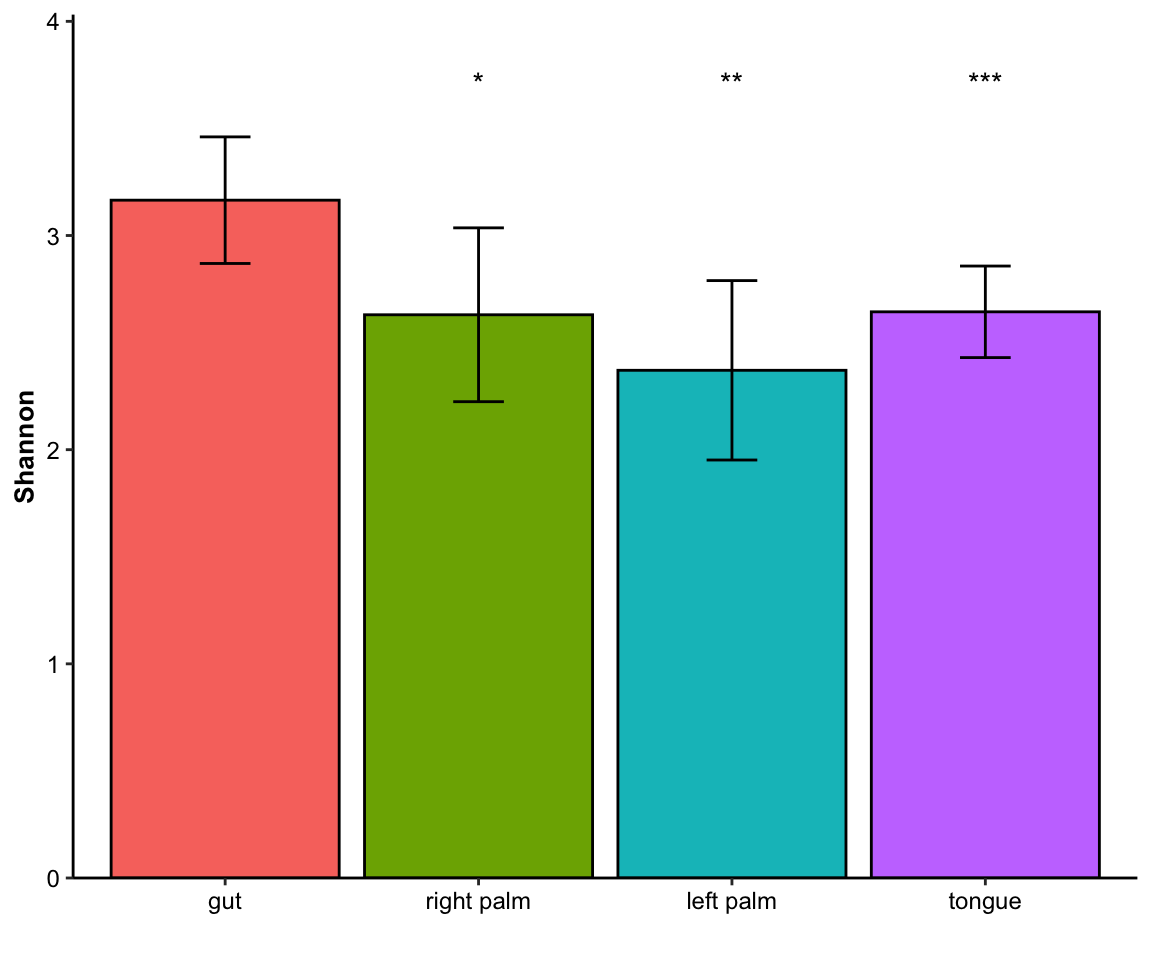
Figure 11.11: barplot(single measure with significant results of ref_group)
- multiple index
plot_barplot(data = dat_alpha,
y_index = c("Shannon", "Chao1", "Observed"),
group = "SampleType",
do_test = TRUE,
method = "wilcox.test")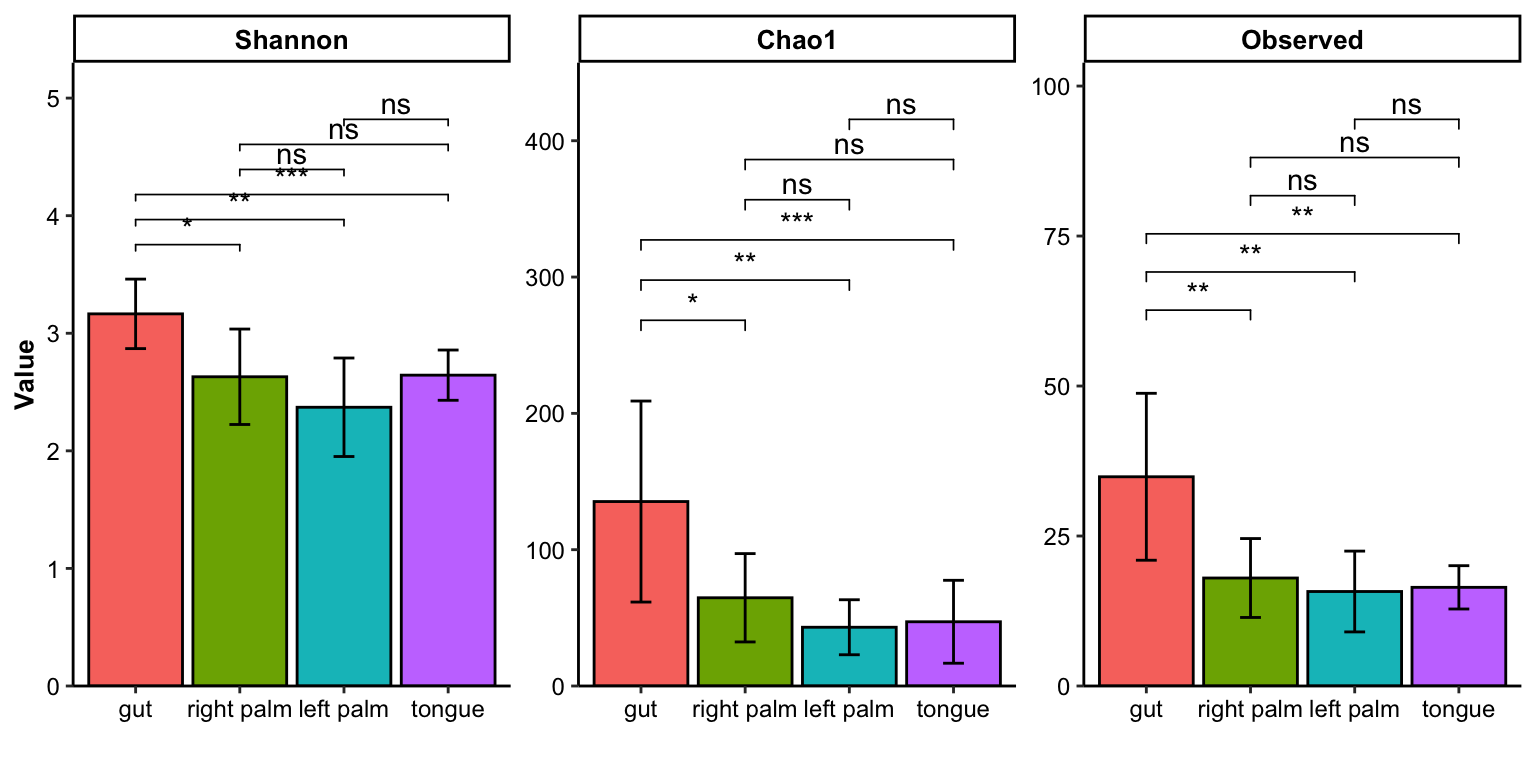
Figure 11.12: barplot(multiple index)
- show group_number in the x-axis break
plot_barplot(data = dat_alpha,
y_index = c("Shannon", "Chao1", "Observed"),
group = "SampleType",
group_names = c("gut", "right palm", "tongue"),
group_color = c("red", "green", "blue"),
do_test = TRUE,
method = "wilcox.test",
group_number = TRUE)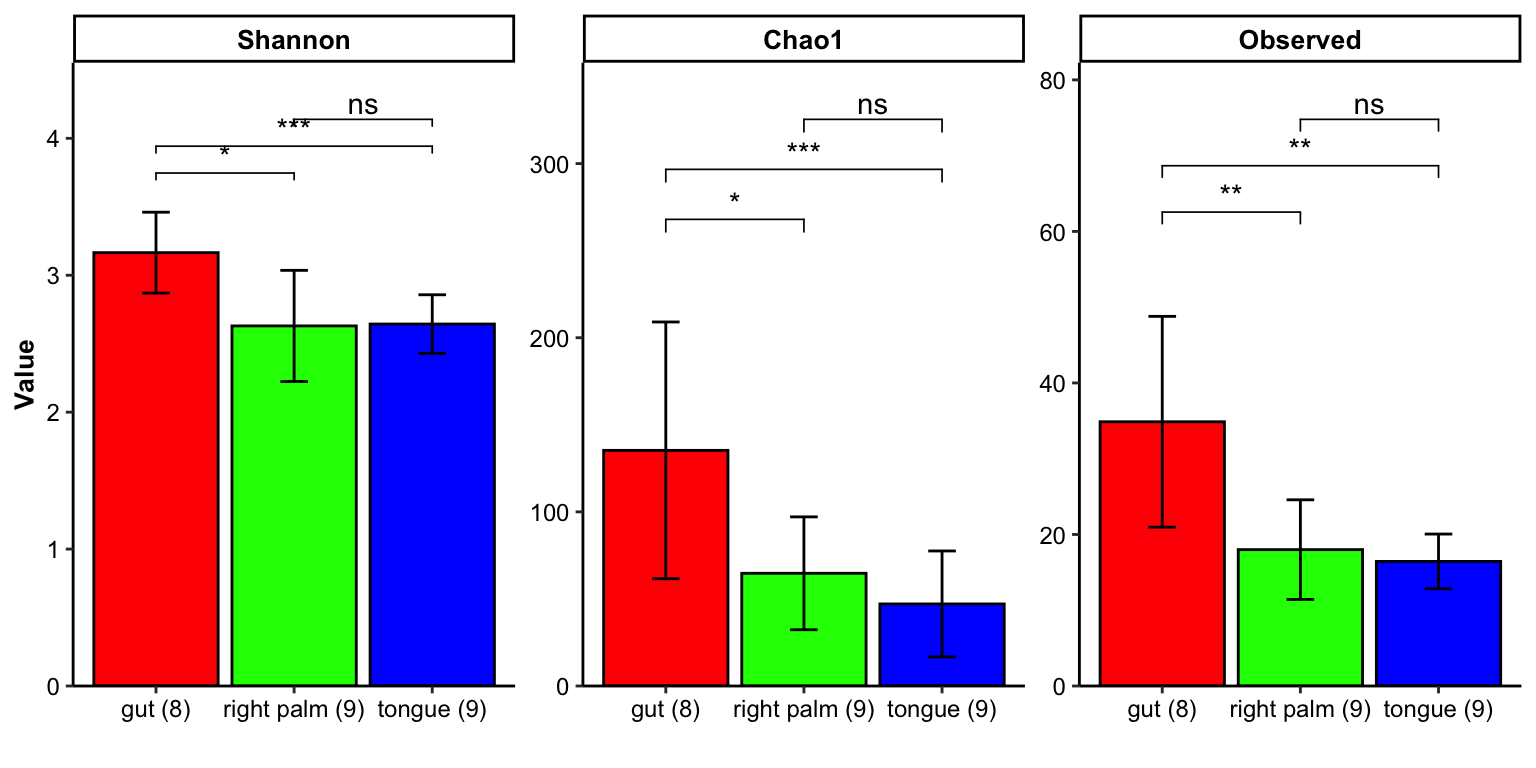
Figure 11.13: barplot(group_number)
11.5 plot_dotplot
plot_dotplot has many parameters, and help you enjoy it.
- single measure
## Bin width defaults to 1/30 of the range of the data. Pick better value with `binwidth`.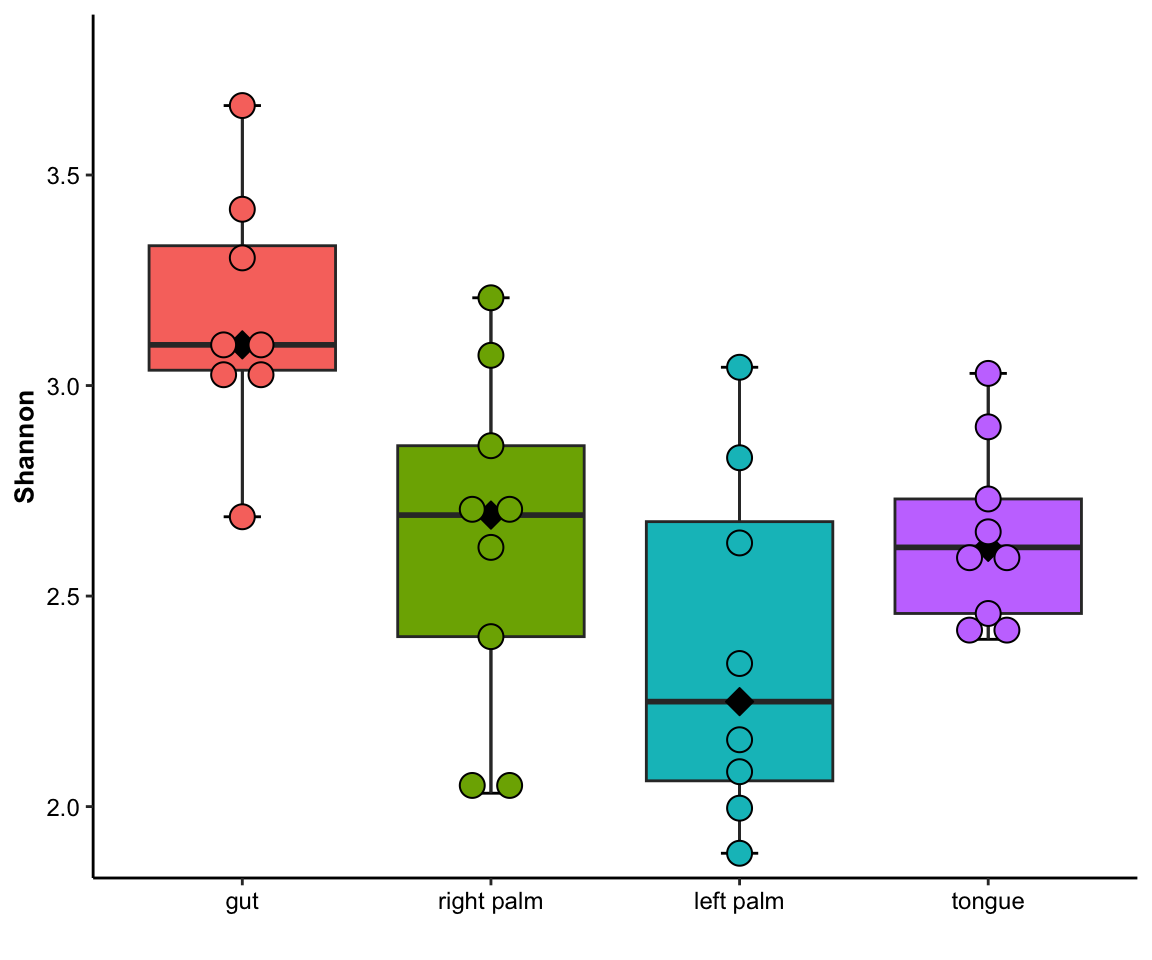
Figure 11.14: dotplot(single measure)
- single measure with significant results
## Bin width defaults to 1/30 of the range of the data. Pick better value with `binwidth`.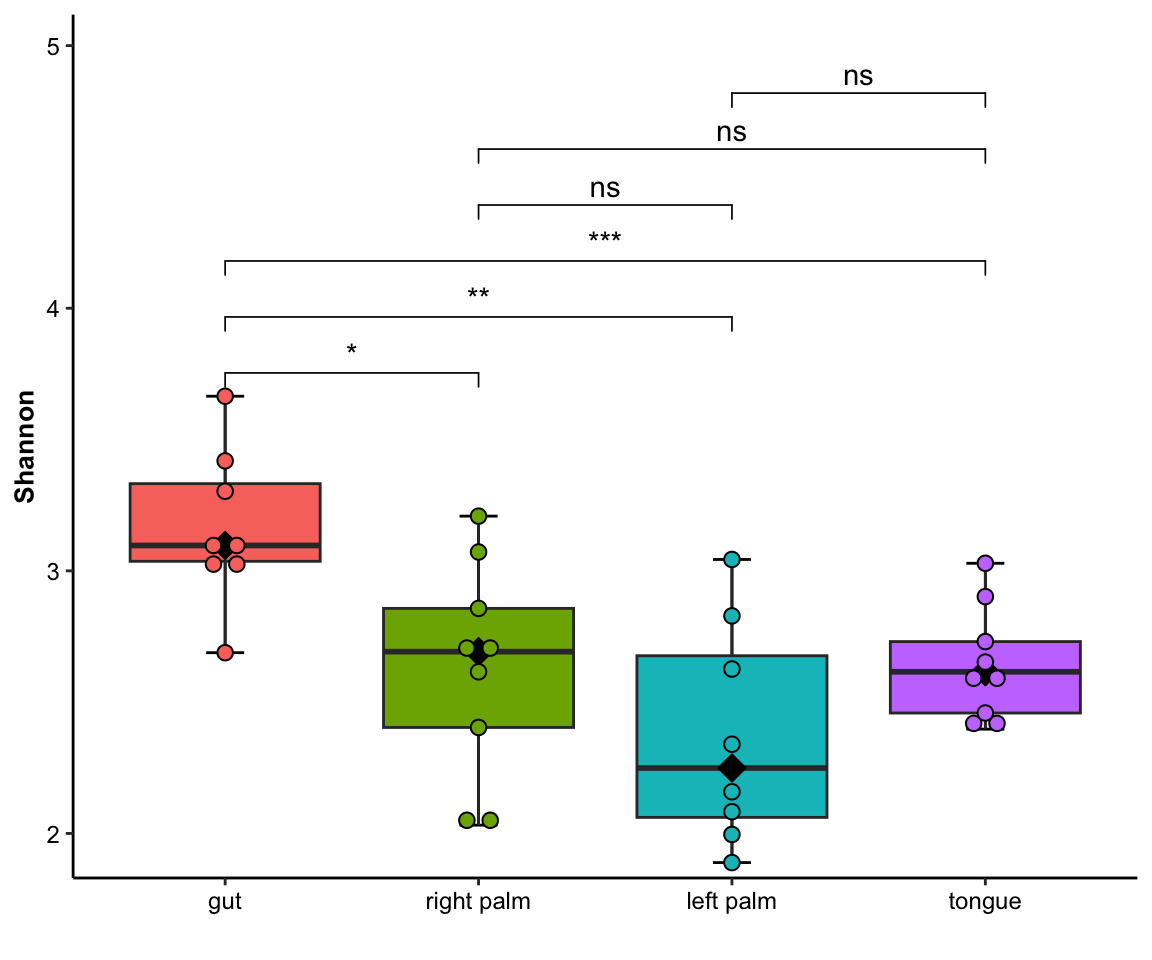
Figure 11.15: dotplot(single measure with significant results)
- single measure with significant results of pairwises
plot_dotplot(data = dat_alpha,
y_index = "Shannon",
group = "SampleType",
do_test = TRUE,
cmp_list = list(c("gut", "right palm"),
c("gut", "tongue")))## Bin width defaults to 1/30 of the range of the data. Pick better value with `binwidth`.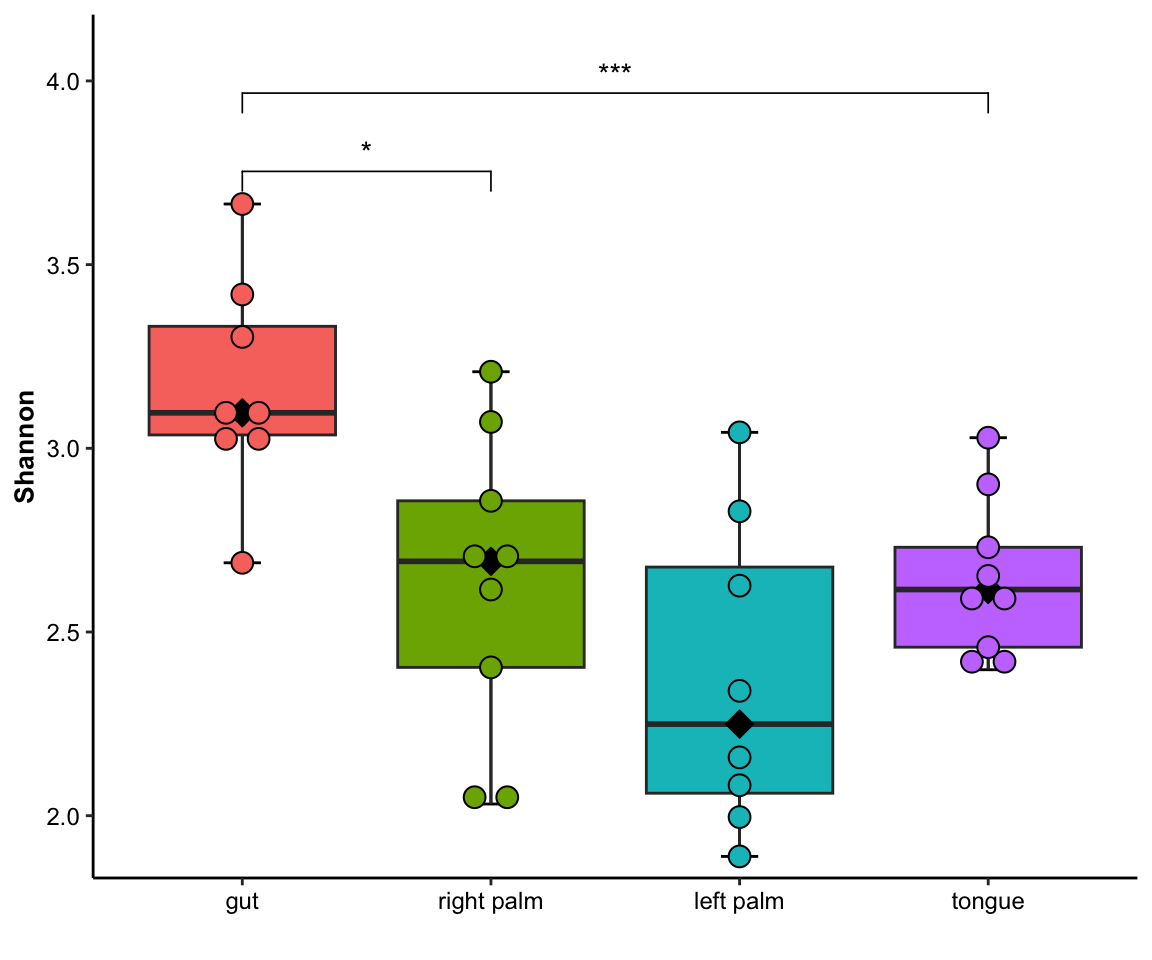
Figure 11.16: dotplot(single measure with significant results of pairwises)
- single measure with significant results of ref_group
plot_dotplot(
data = dat_alpha,
y_index = "Shannon",
group = "SampleType",
do_test = TRUE,
ref_group = "gut")## Bin width defaults to 1/30 of the range of the data. Pick better value with `binwidth`.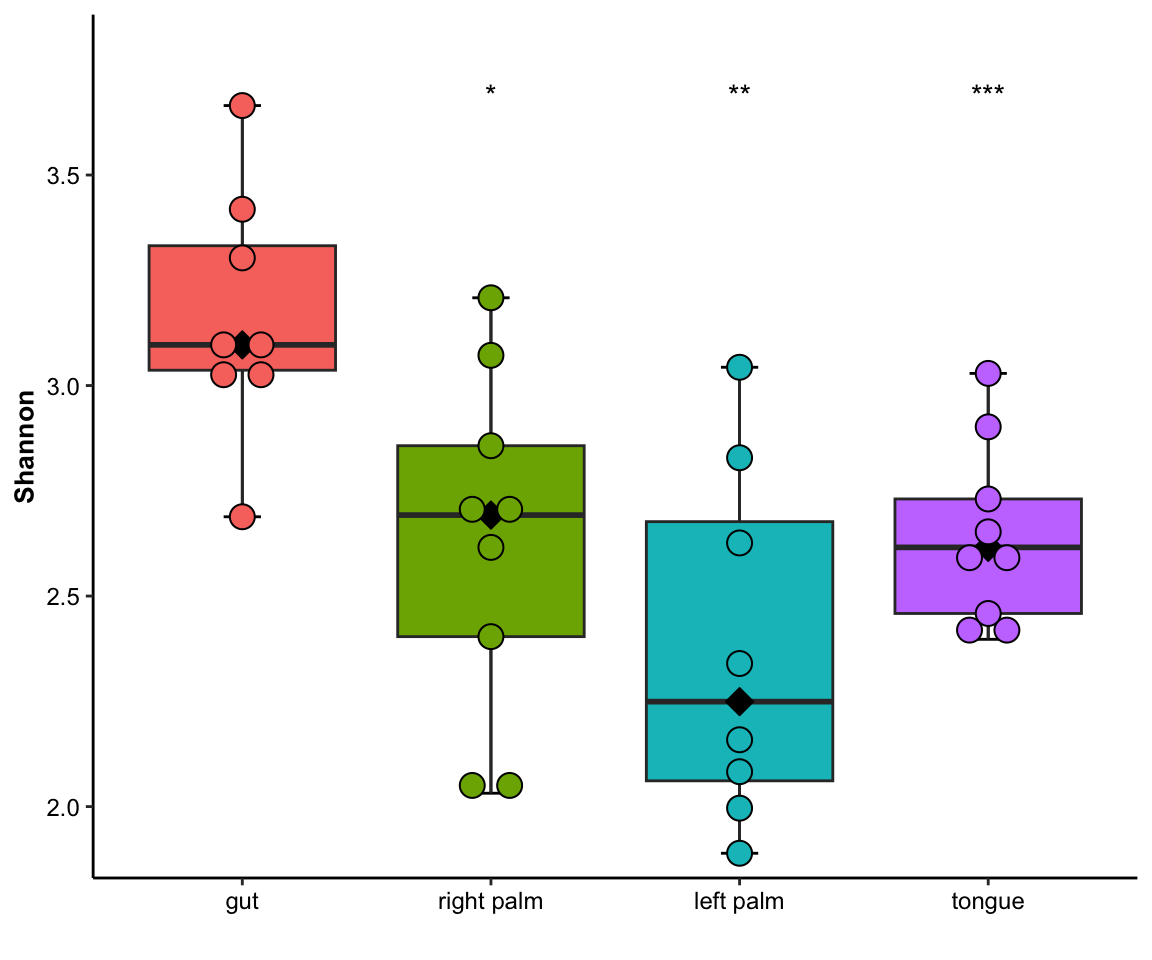
Figure 11.17: dotplot(single measure with significant results of ref_group)
- dot size and median size
plot_dotplot(
data = dat_alpha,
y_index = "Shannon",
group = "SampleType",
do_test = TRUE,
ref_group = "gut",
dotsize = 0.5,
mediansize = 2)## Bin width defaults to 1/30 of the range of the data. Pick better value with `binwidth`.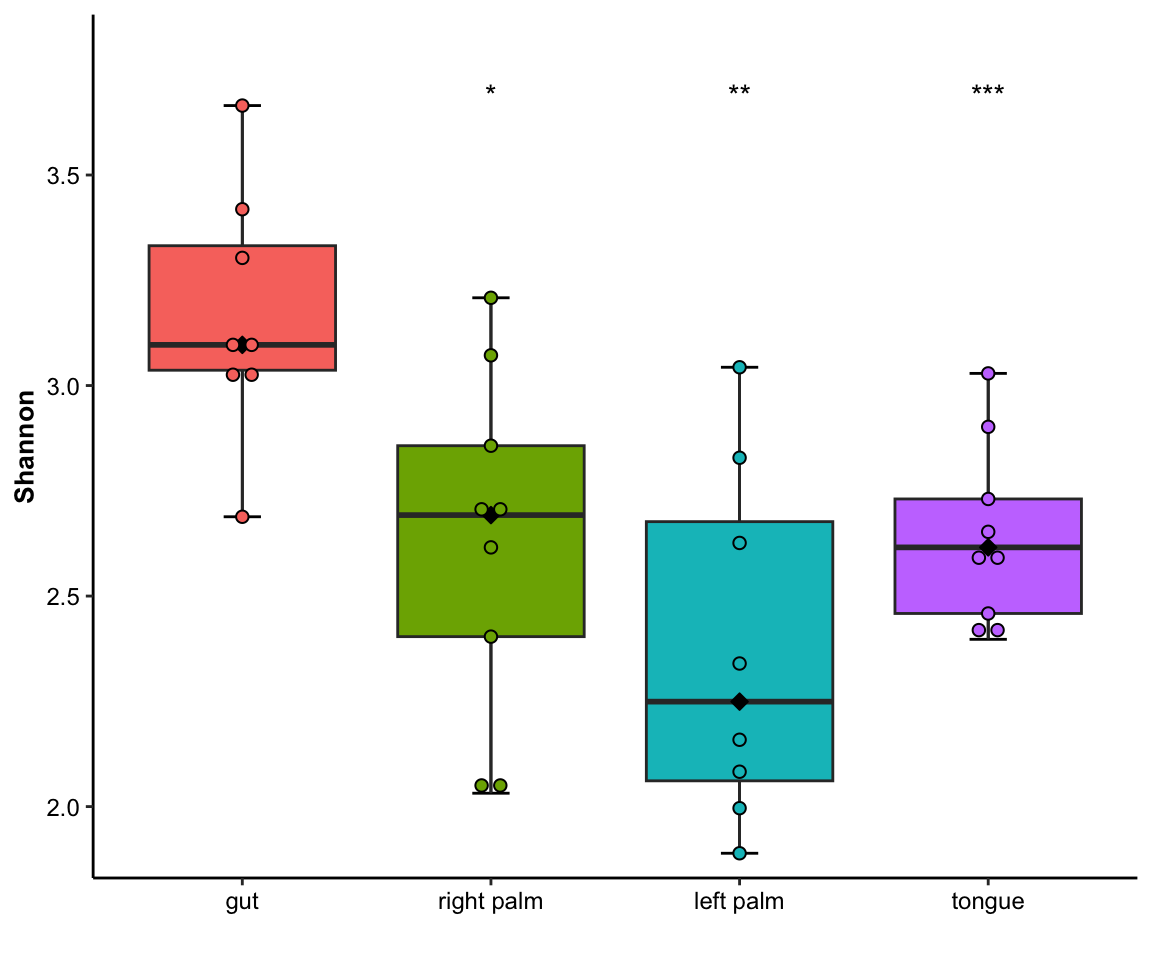
Figure 11.18: dotplot(dot size and median size)
- multiple index
plot_dotplot(
data = dat_alpha,
y_index = c("Shannon", "Chao1", "Observed"),
group = "SampleType",
do_test = TRUE,
method = "wilcox.test")## Bin width defaults to 1/30 of the range of the data. Pick better value with `binwidth`.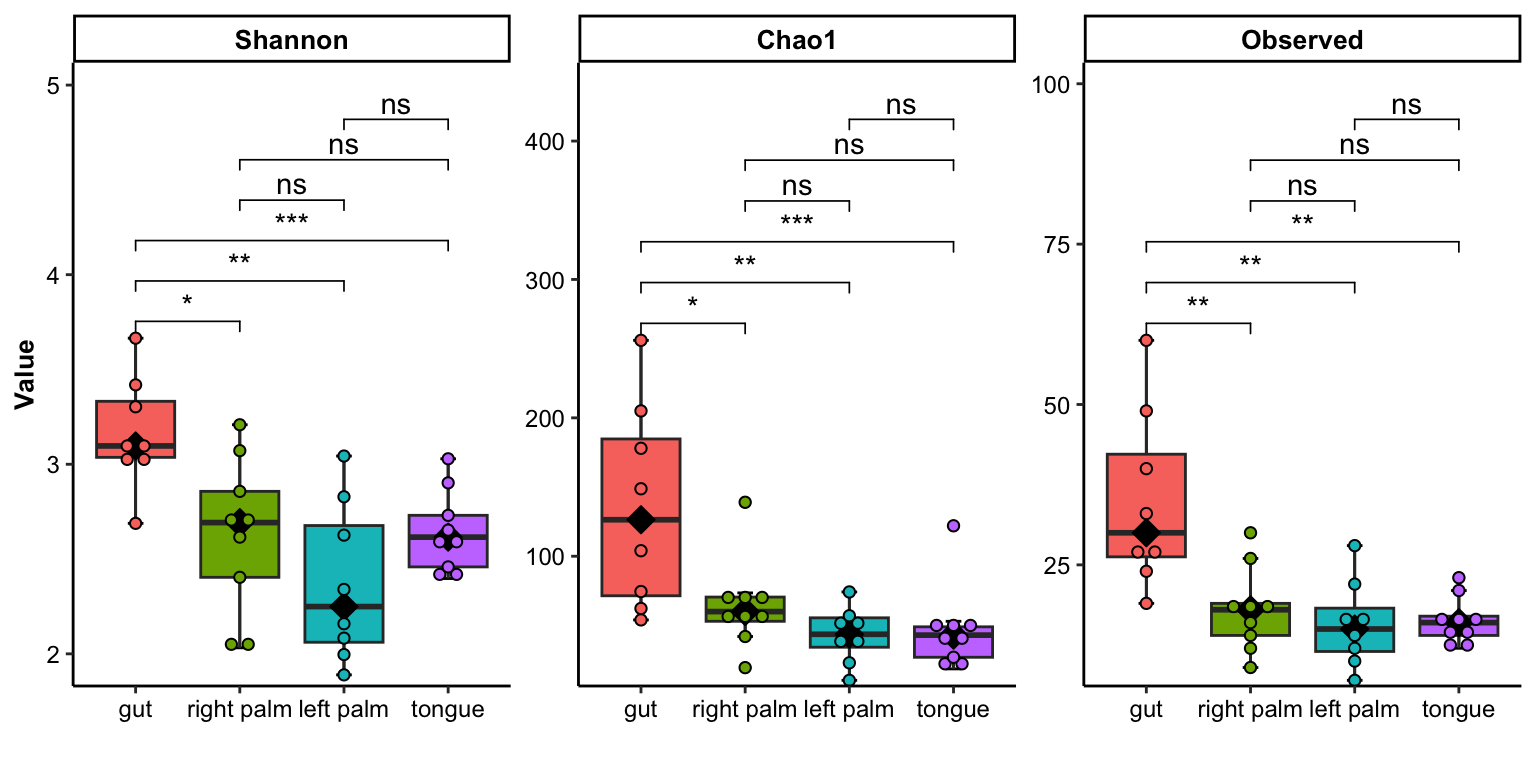
Figure 11.19: dotplot(multiple index)
- multiple index with errorbar
plot_dotplot(
data = dat_alpha,
y_index = c("Shannon", "Chao1", "Observed"),
group = "SampleType",
do_test = TRUE,
show_type = "errorbar",
method = "wilcox.test")## Bin width defaults to 1/30 of the range of the data. Pick better value with `binwidth`.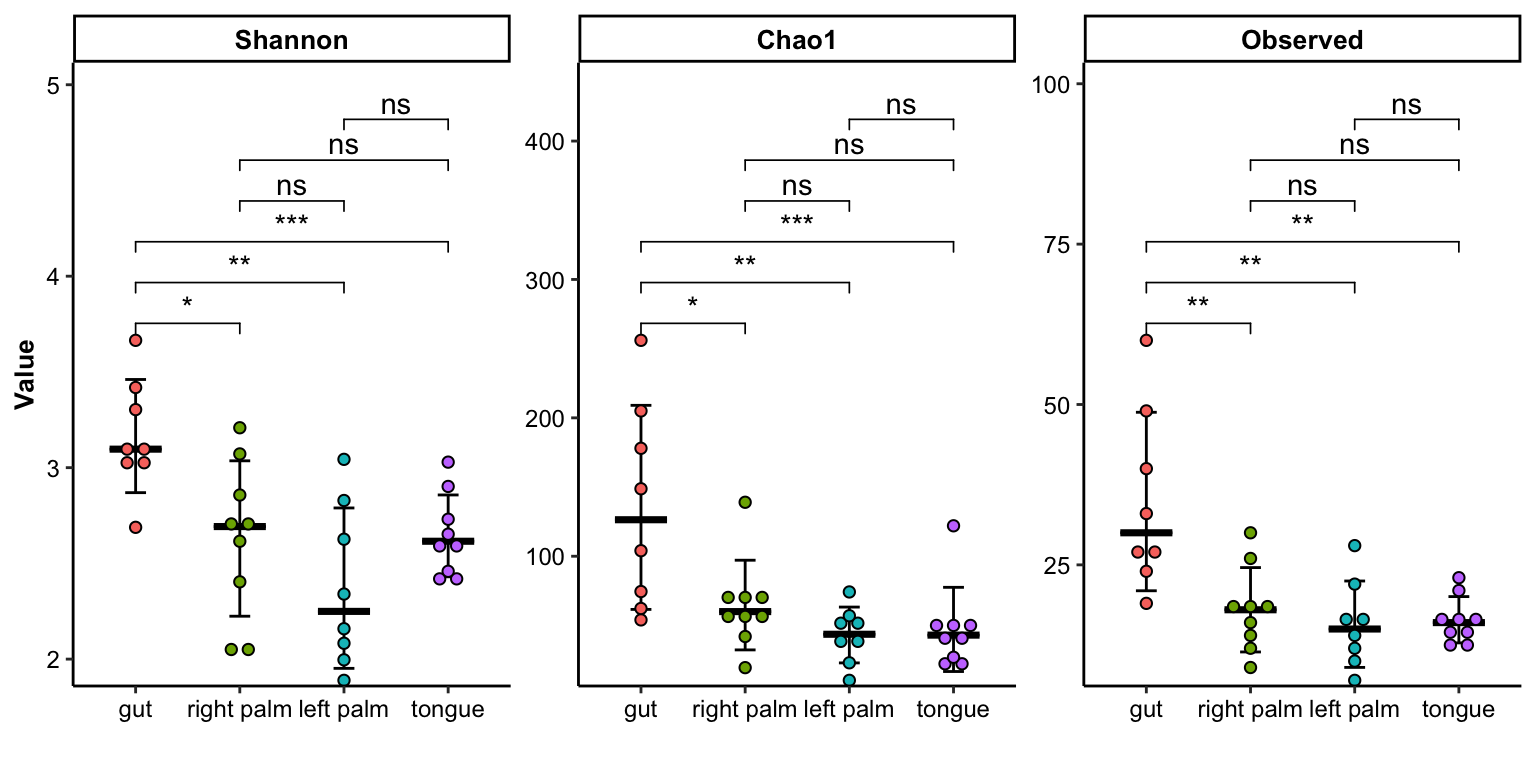
Figure 11.20: dotplot(multiple index errorbar)
- show group_number in the x-axis break
plot_dotplot(data = dat_alpha,
y_index = c("Shannon", "Chao1", "Observed"),
group = "SampleType",
group_names = c("gut", "right palm", "tongue"),
group_color = c("red", "green", "blue"),
show_type = "errorbar",
do_test = TRUE,
method = "wilcox.test",
group_number = TRUE)## Bin width defaults to 1/30 of the range of the data. Pick better value with `binwidth`.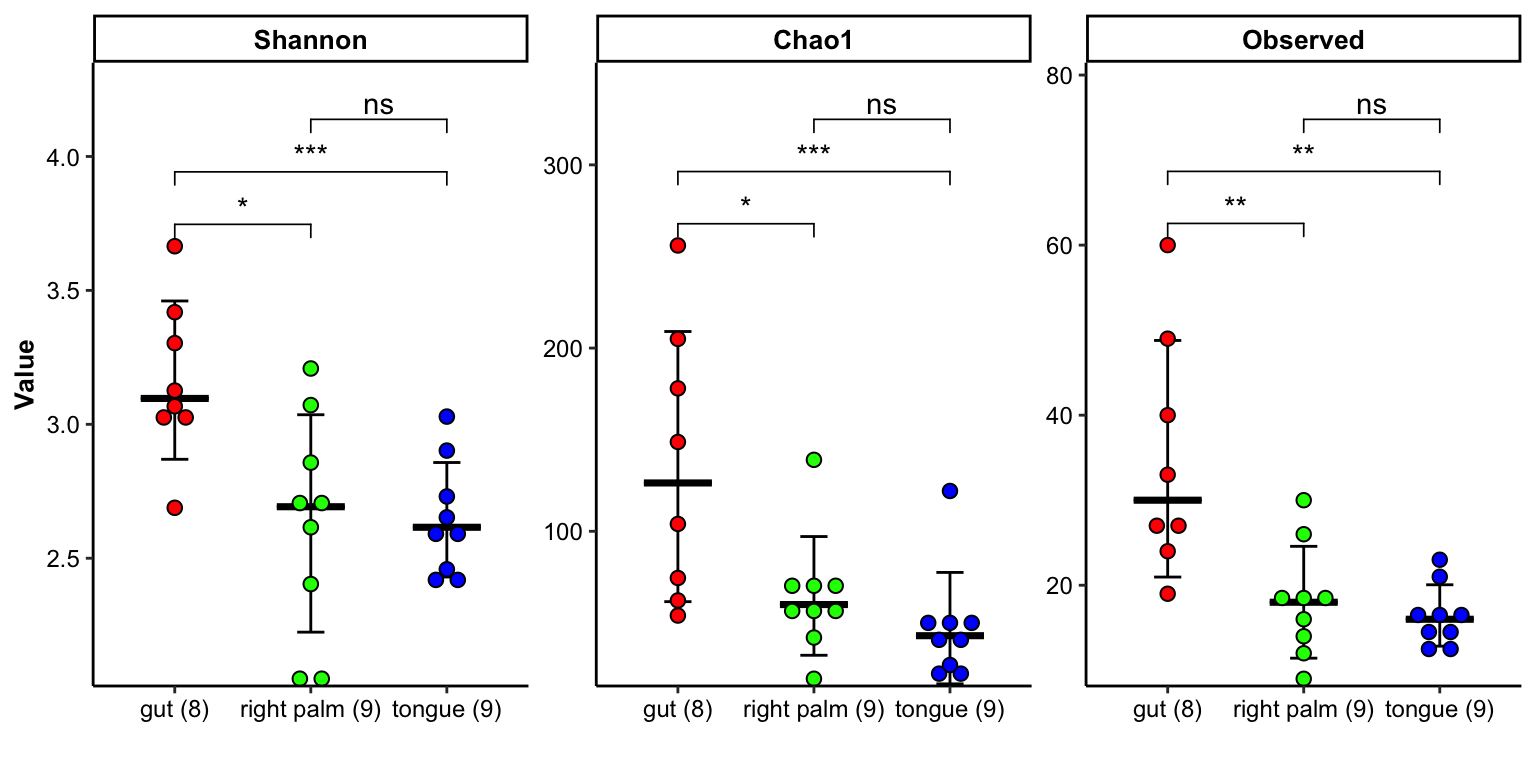
Figure 11.21: dotplot(group_number)
11.6 plot_correlation_boxplot
Help you enjoy plot_correlation_boxplot.
plot_correlation_boxplot(
data = dat_alpha,
x_index = "Chao1",
y_index = "Shannon",
group = "SampleType")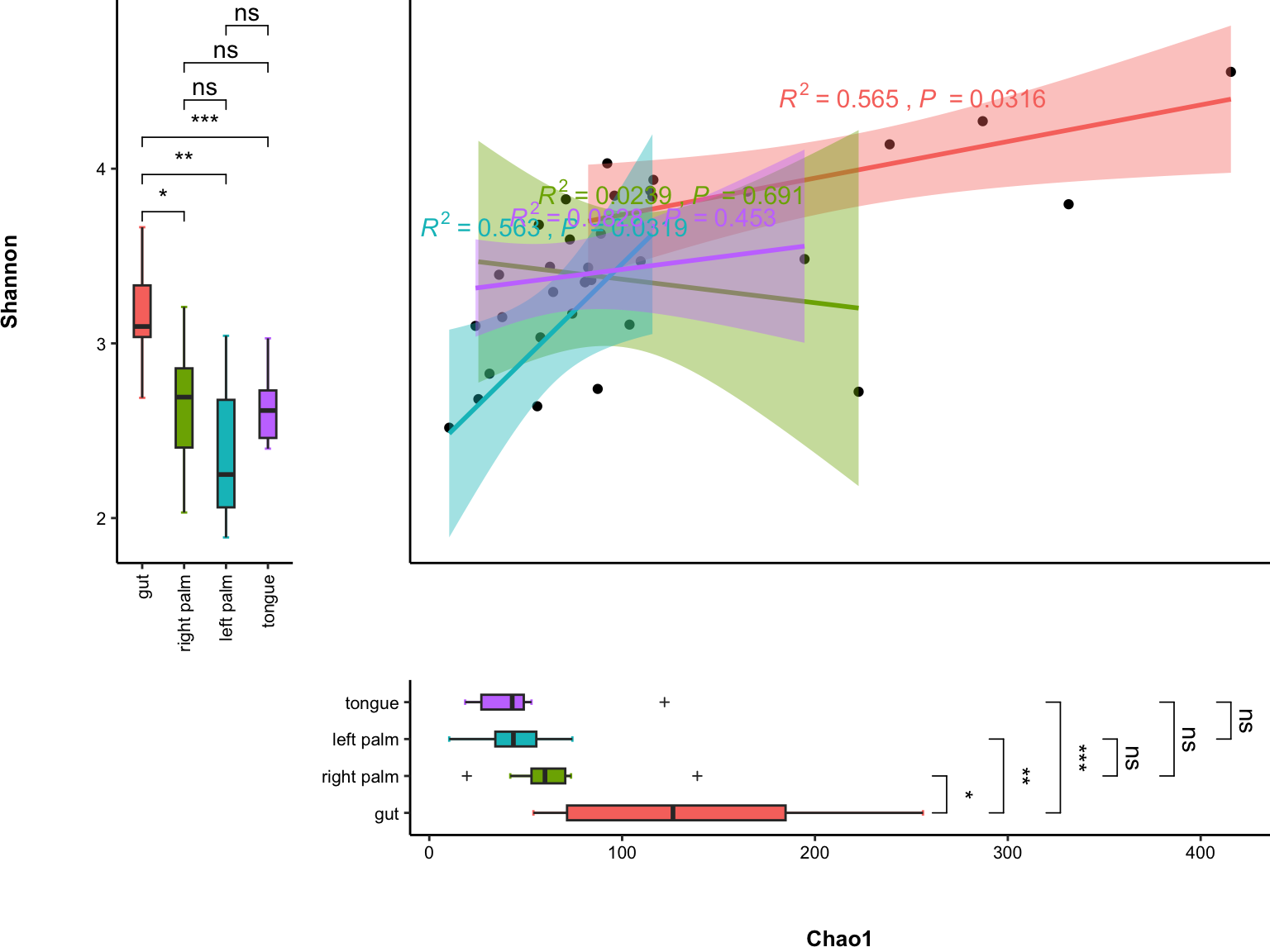
Figure 11.22: correlation with boxplot
11.7 plot_correlation_density
Help you enjoy plot_correlation_density.
plot_correlation_density(
data = dat_alpha,
x_index = "Chao1",
y_index = "Shannon",
group = "SampleType")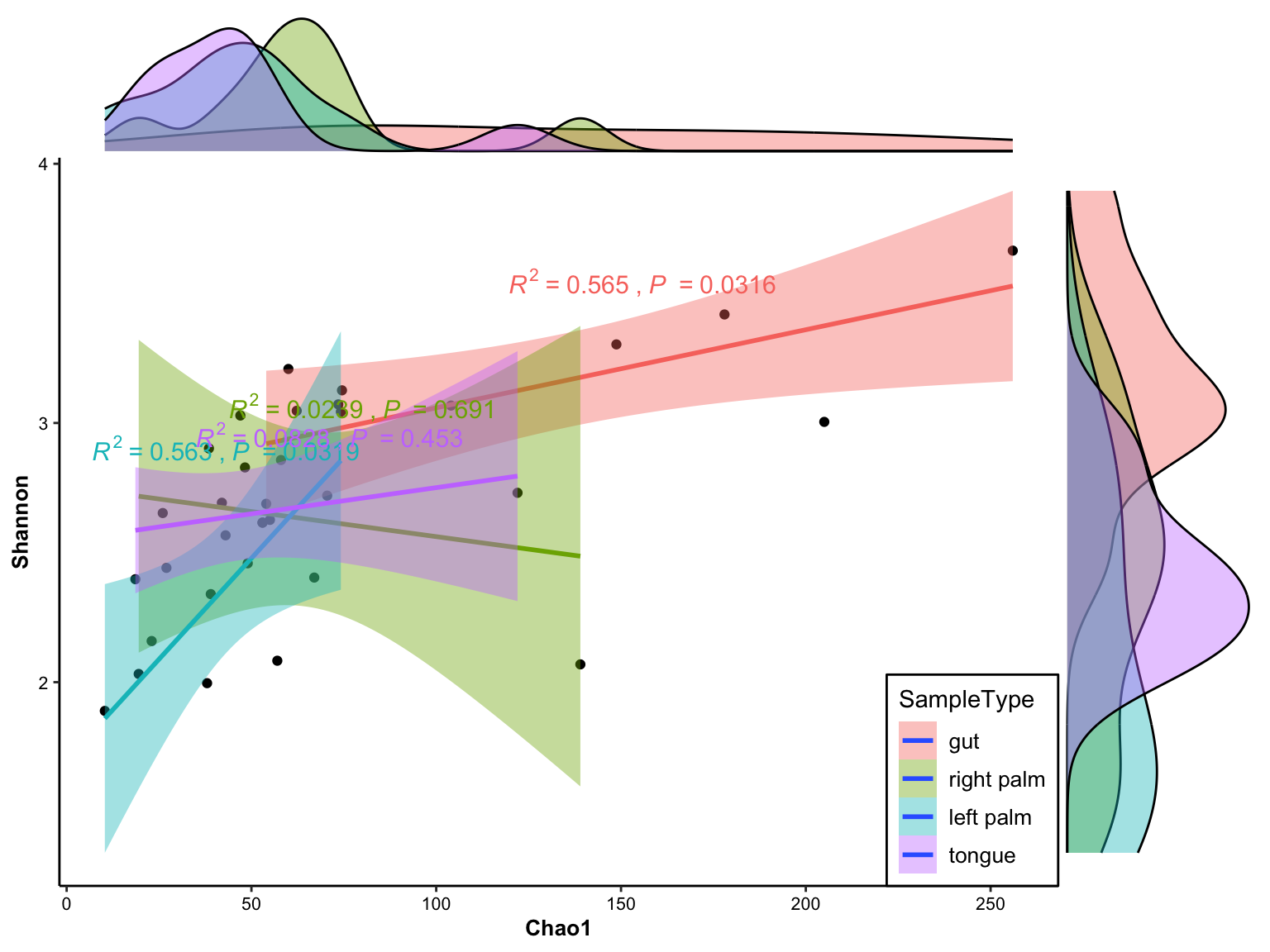
Figure 11.23: correlation with density
11.8 plot_Ordination
plot_Ordination provides too many parameters for users to display the ordination results by using ggplot2 format. Here is the ordinary pattern.
data("dada2_ps")
# step1: Removing samples of specific group in phyloseq-class object
dada2_ps_remove_BRS <- get_GroupPhyloseq(
ps = dada2_ps,
group = "Group",
group_names = "QC")
# step2: Rarefying counts in phyloseq-class object
dada2_ps_rarefy <- norm_rarefy(object = dada2_ps_remove_BRS,
size = 51181)
# step3: Extracting specific taxa phyloseq-class object
dada2_ps_rare_genus <- summarize_taxa(ps = dada2_ps_rarefy,
taxa_level = "Genus",
absolute = TRUE)
# step4: Aggregating low relative abundance or unclassified taxa into others
# dada2_ps_genus_LRA <- summarize_LowAbundance_taxa(ps = dada2_ps_rare_genus,
# cutoff = 10,
# unclass = TRUE)
# step4: Filtering the low relative abundance or unclassified taxa by the threshold
dada2_ps_genus_filter <- run_filter(ps = dada2_ps_rare_genus,
cutoff = 10,
unclass = TRUE)
# step5: Trimming the taxa with low occurrence less than threshold
dada2_ps_genus_filter_trim <- run_trim(object = dada2_ps_genus_filter,
cutoff = 0.2,
trim = "feature")
dada2_ps_genus_filter_trim## phyloseq-class experiment-level object
## otu_table() OTU Table: [ 83 taxa and 23 samples ]
## sample_data() Sample Data: [ 23 samples by 1 sample variables ]
## tax_table() Taxonomy Table: [ 83 taxa by 6 taxonomic ranks ]ordination_PCA <- run_ordination(
ps = dada2_ps_genus_filter_trim,
group = "Group",
method = "PCA")
names(ordination_PCA)## [1] "fit" "dat" "explains" "eigvalue" "PERMANOVA" "BETADISPER" "axis_taxa_cor"- Ordinary pattern
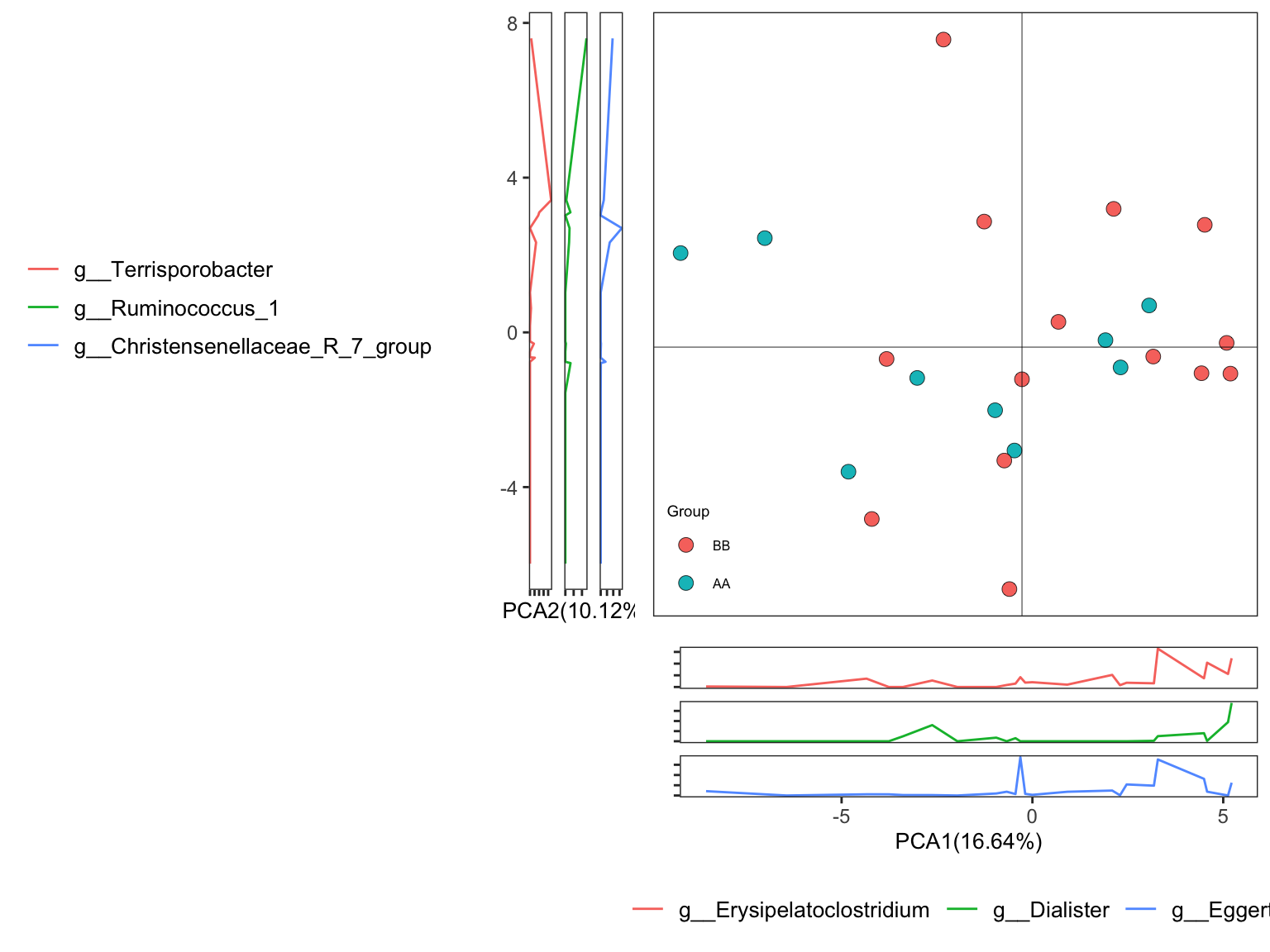
Figure 11.24: plot_Ordination (Ordinary pattern)
- plot with SampleID and setting group colors
plot_Ordination(ResultList = ordination_PCA,
group = "Group",
group_names = c("AA", "BB"),
group_color = c("blue", "red"),
circle_type = "ellipse",
sample = TRUE)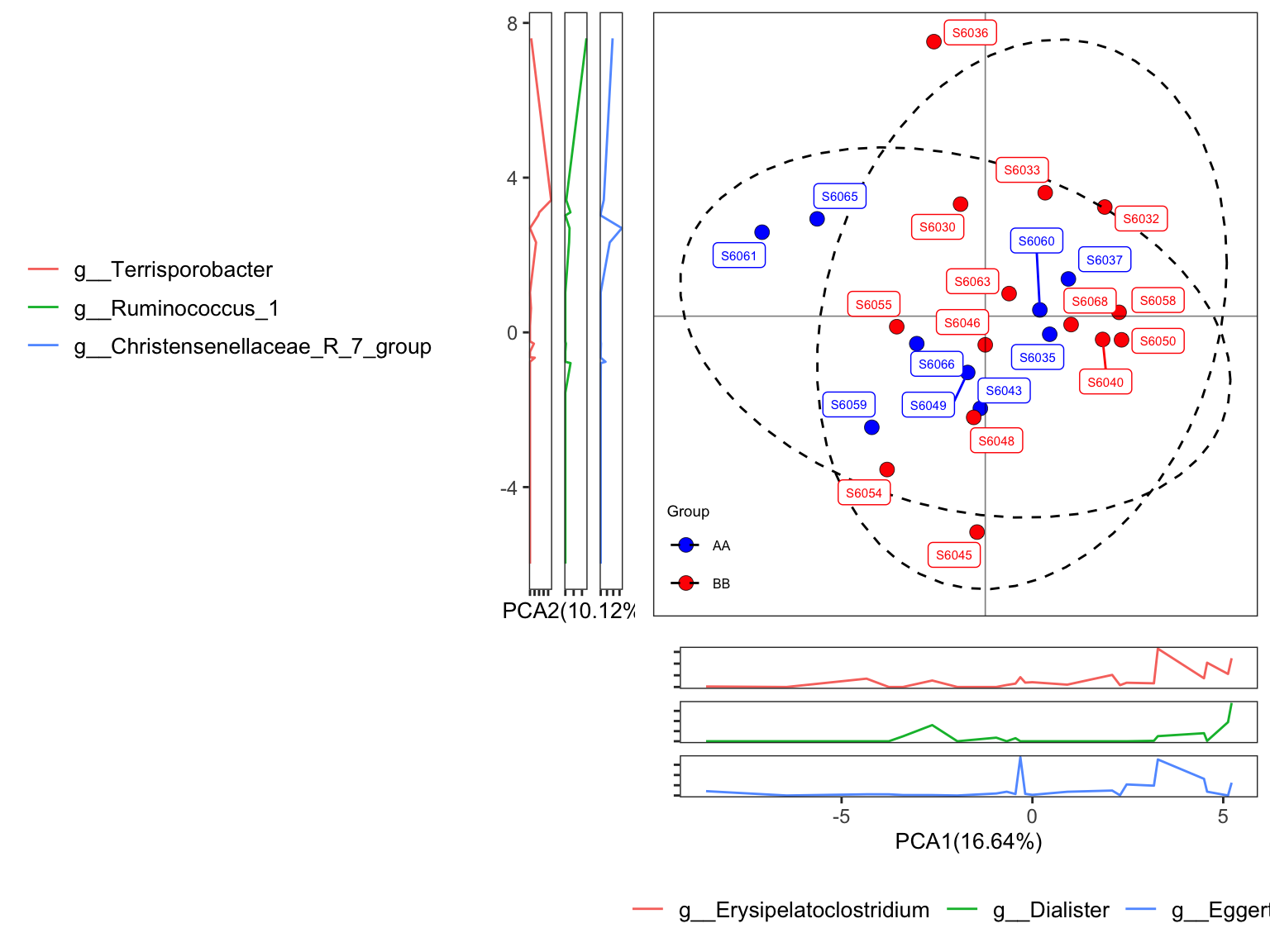
Figure 11.25: plot_Ordination (Ordinary pattern with SampleID)
- ellipse with 95% confidence interval
plot_Ordination(ResultList = ordination_PCA,
group = "Group",
group_names = c("AA", "BB"),
group_color = c("blue", "red"),
circle_type = "ellipse_CI",
sample = TRUE)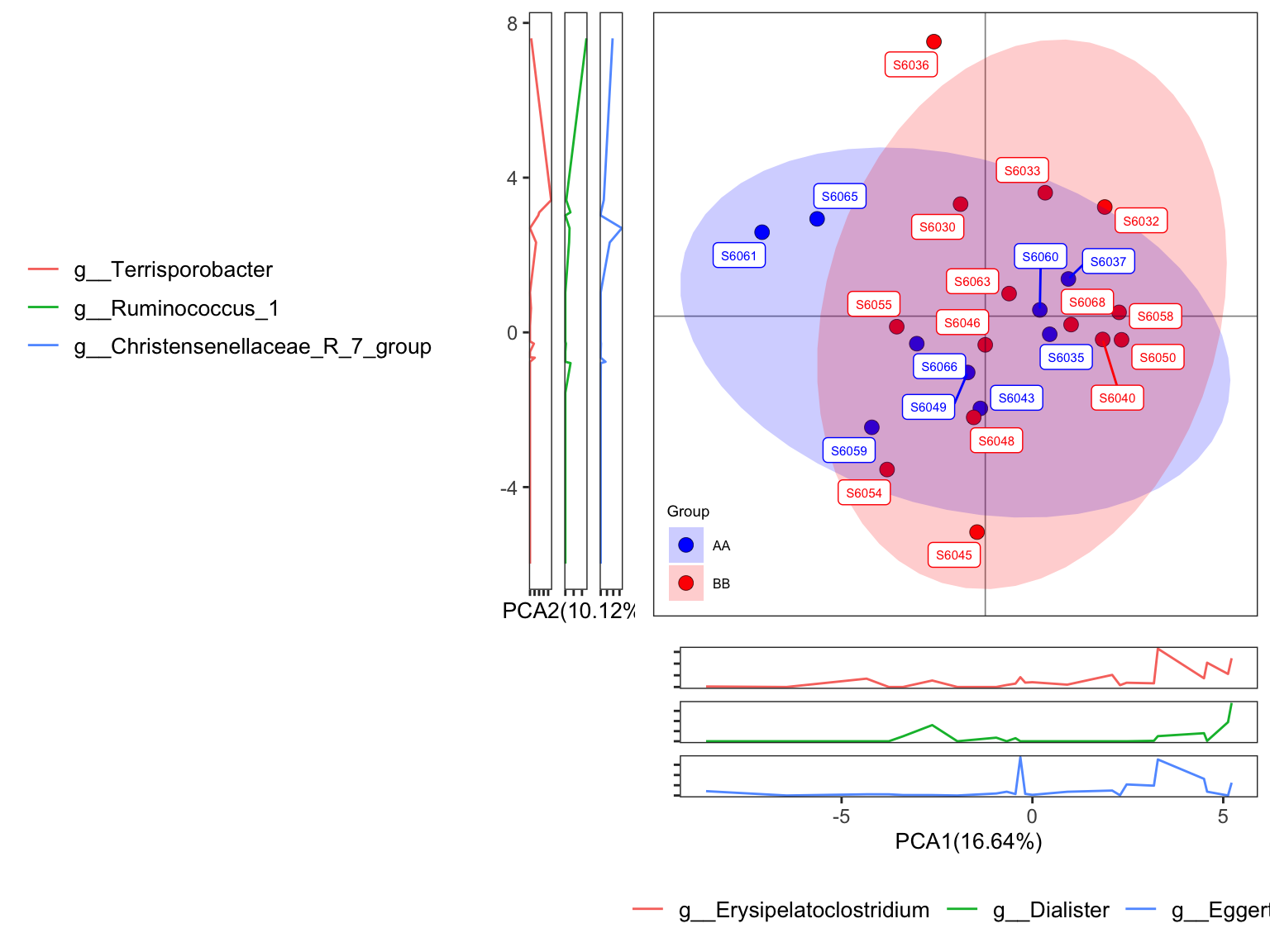
Figure 11.26: plot_Ordination (ellipse with 95% confidence interval)
- ellipse with groups
plot_Ordination(ResultList = ordination_PCA,
group = "Group",
group_names = c("AA", "BB"),
group_color = c("blue", "red"),
circle_type = "ellipse_groups",
sample = TRUE)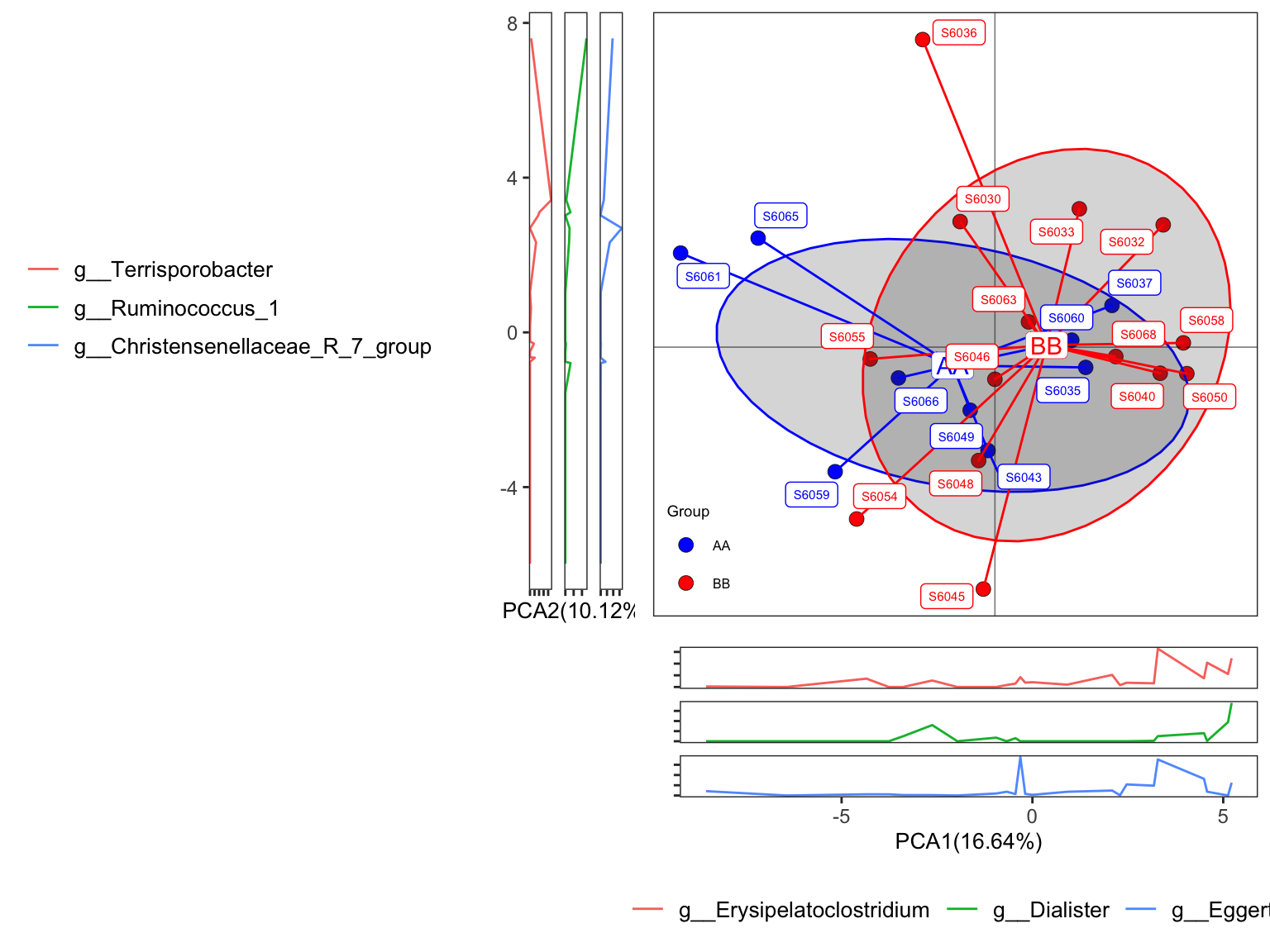
Figure 11.27: plot_Ordination (ellipse with groups)
- ellipse with border line
plot_Ordination(ResultList = ordination_PCA,
group = "Group",
group_names = c("AA", "BB"),
group_color = c("blue", "red"),
circle_type = "ellipse_line",
sample = TRUE)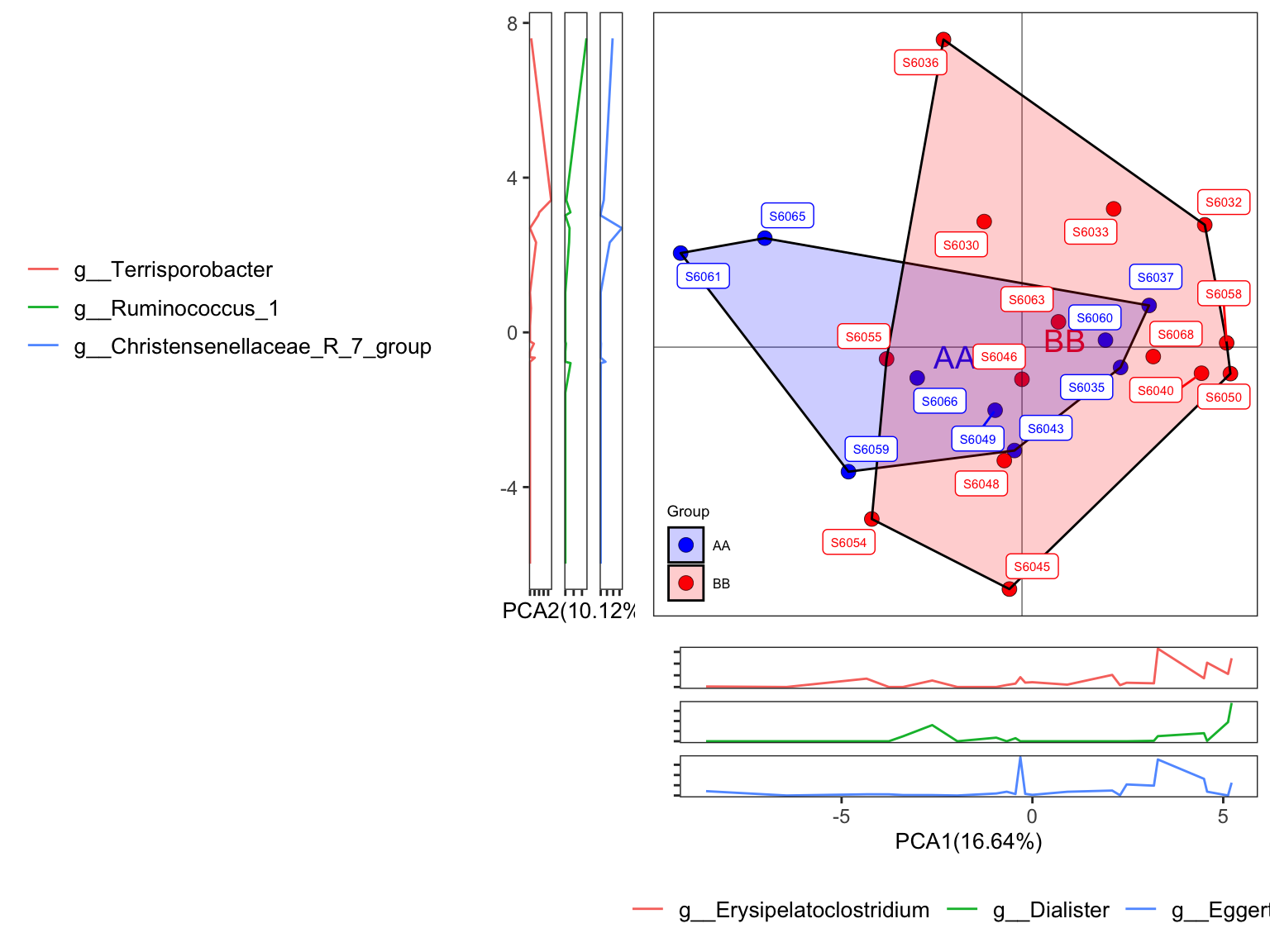
Figure 11.28: plot_Ordination (ellipse with border line)
- plot with SampleID and sideboxplot and setting group colors
plot_Ordination(ResultList = ordination_PCA,
group = "Group",
group_names = c("AA", "BB"),
group_color = c("blue", "red"),
circle_type = "ellipse",
sidelinechart = FALSE,
sideboxplot = TRUE,
sample = TRUE)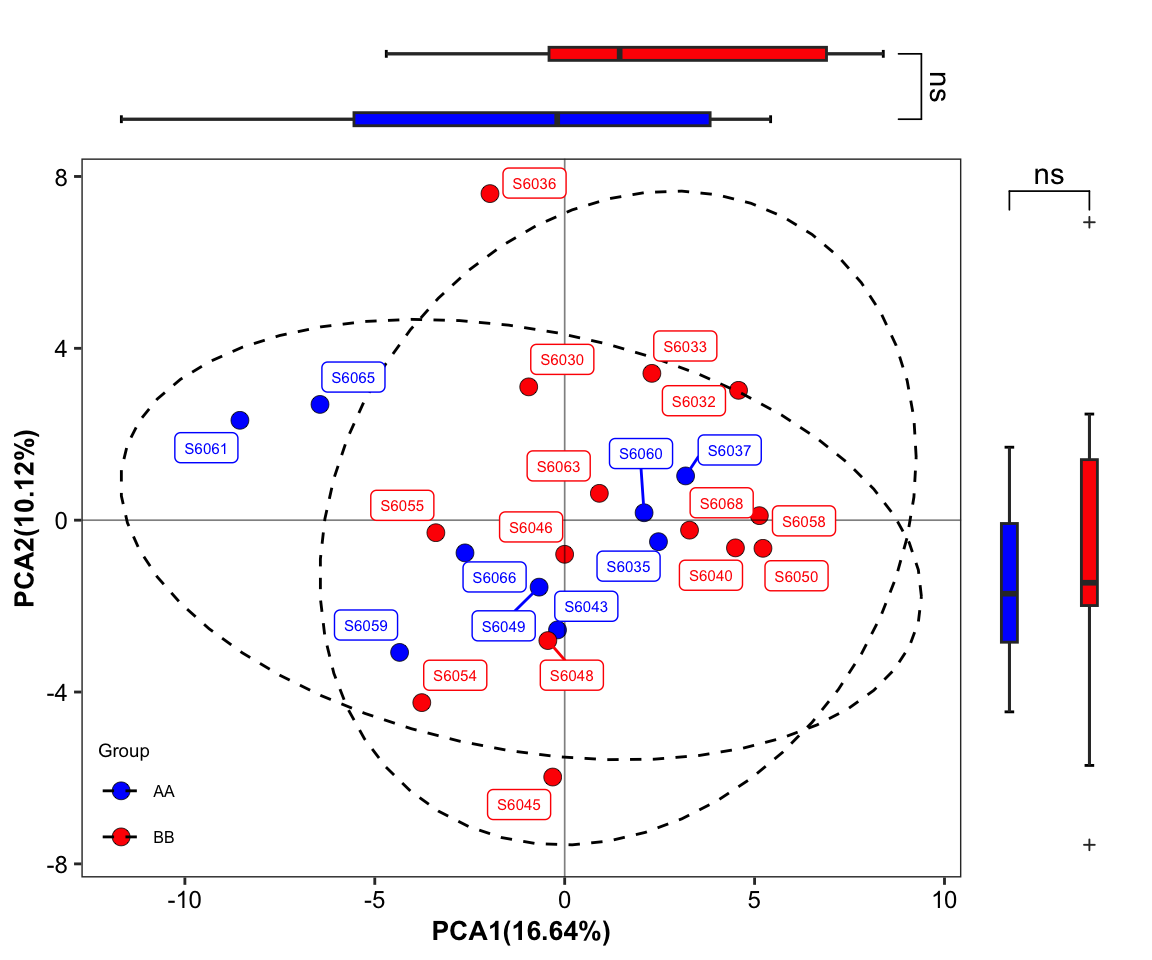
Figure 11.29: plot_Ordination (Ordinary pattern with SampleID sideboxplot)
- plot with SampleID and sideboxplot and setting group colors 2
plot_Ordination(ResultList = ordination_PCA,
group = "Group",
group_names = c("AA", "BB"),
group_color = c("blue", "red"),
circle_type = "ellipse_CI",
sidelinechart = FALSE,
sideboxplot = TRUE,
sample = TRUE)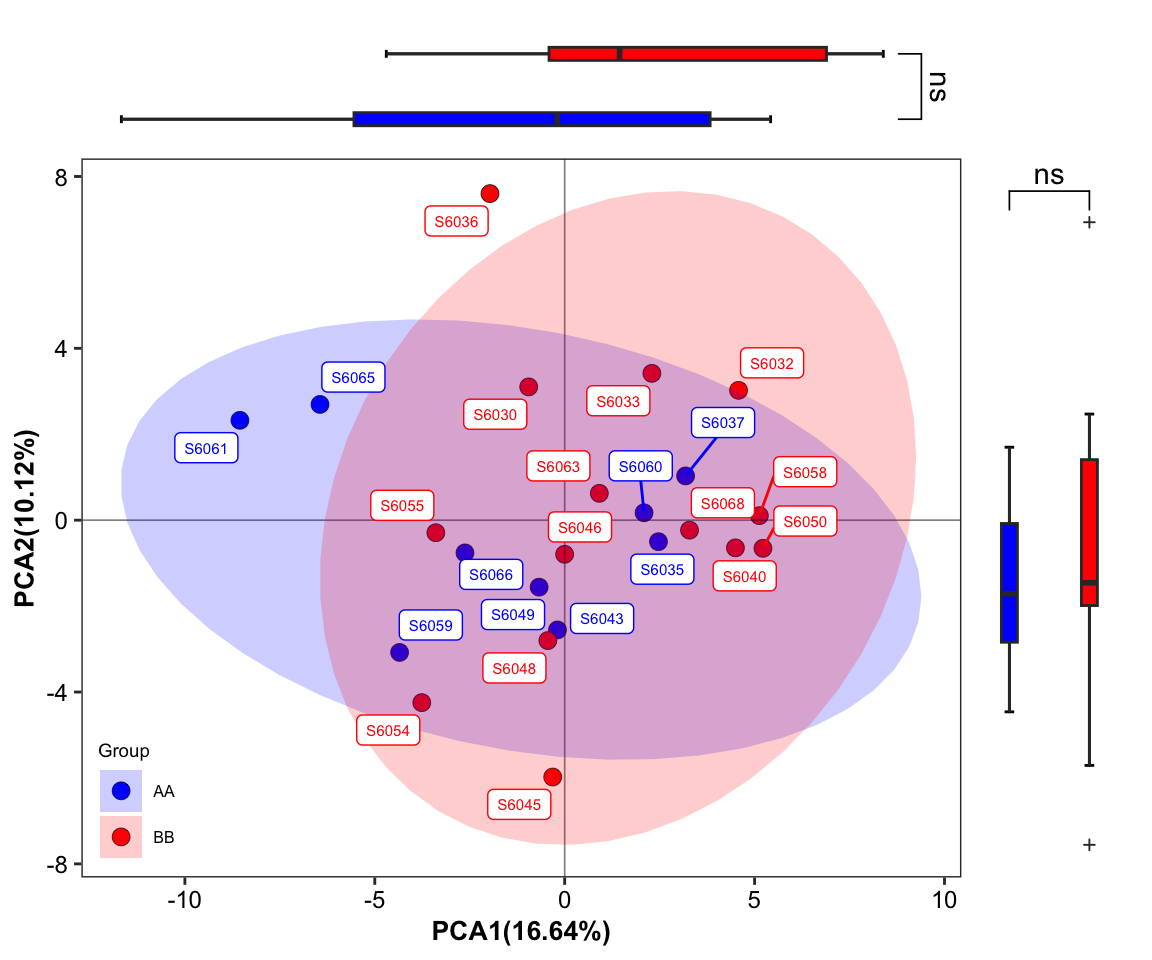
Figure 11.30: plot_Ordination (ellipse_CI with SampleID sideboxplot)
- plot with SampleID and sideboxplot and setting group colors 3
plot_Ordination(ResultList = ordination_PCA,
group = "Group",
group_names = c("AA", "BB"),
group_color = c("blue", "red"),
circle_type = "ellipse_groups",
sidelinechart = FALSE,
sideboxplot = TRUE,
sample = TRUE)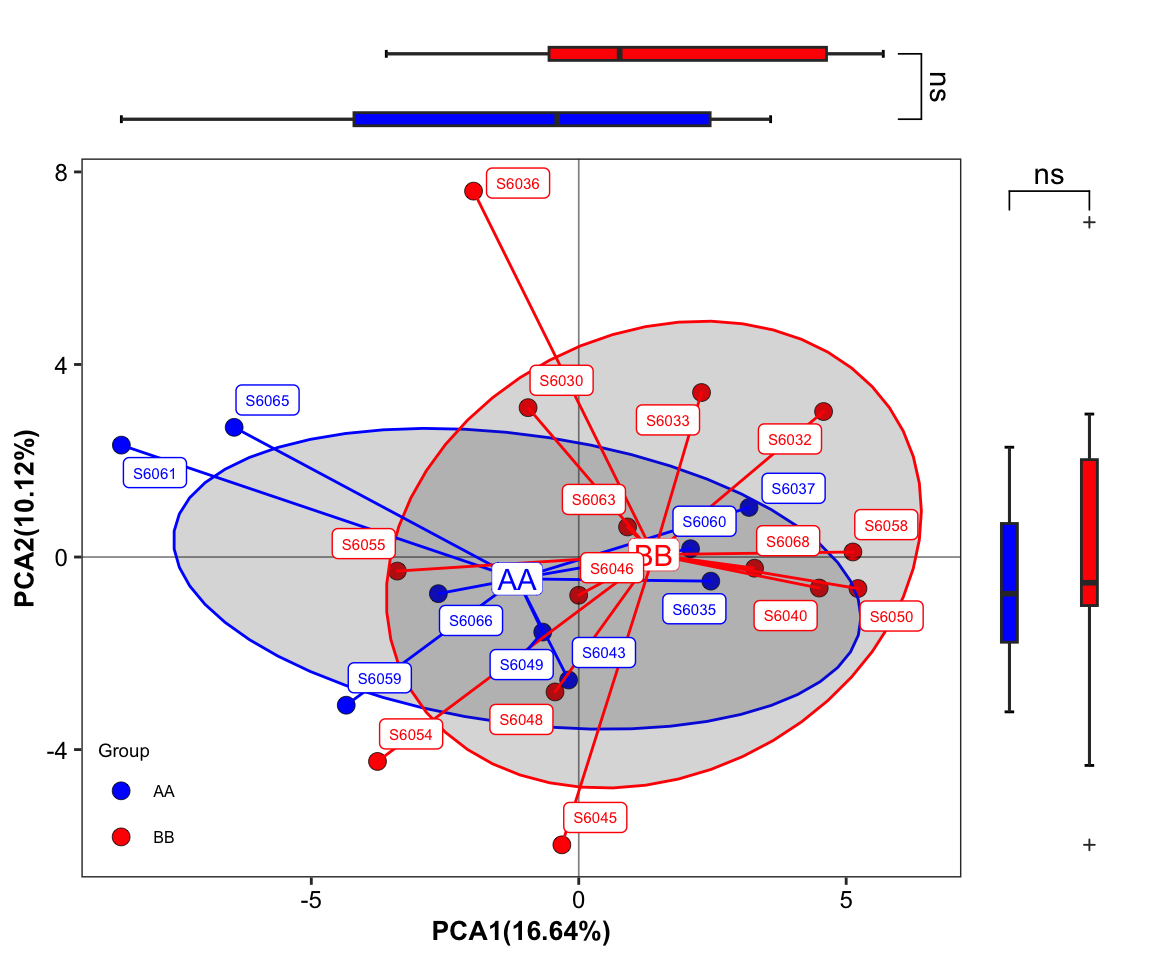
Figure 11.31: plot_Ordination (ellipse_groups with SampleID sideboxplot)
- plot with SampleID and sideboxplot and setting group colors 4
plot_Ordination(ResultList = ordination_PCA,
group = "Group",
group_names = c("AA", "BB"),
group_color = c("blue", "red"),
circle_type = "ellipse_line",
sidelinechart = FALSE,
sideboxplot = TRUE,
sample = TRUE)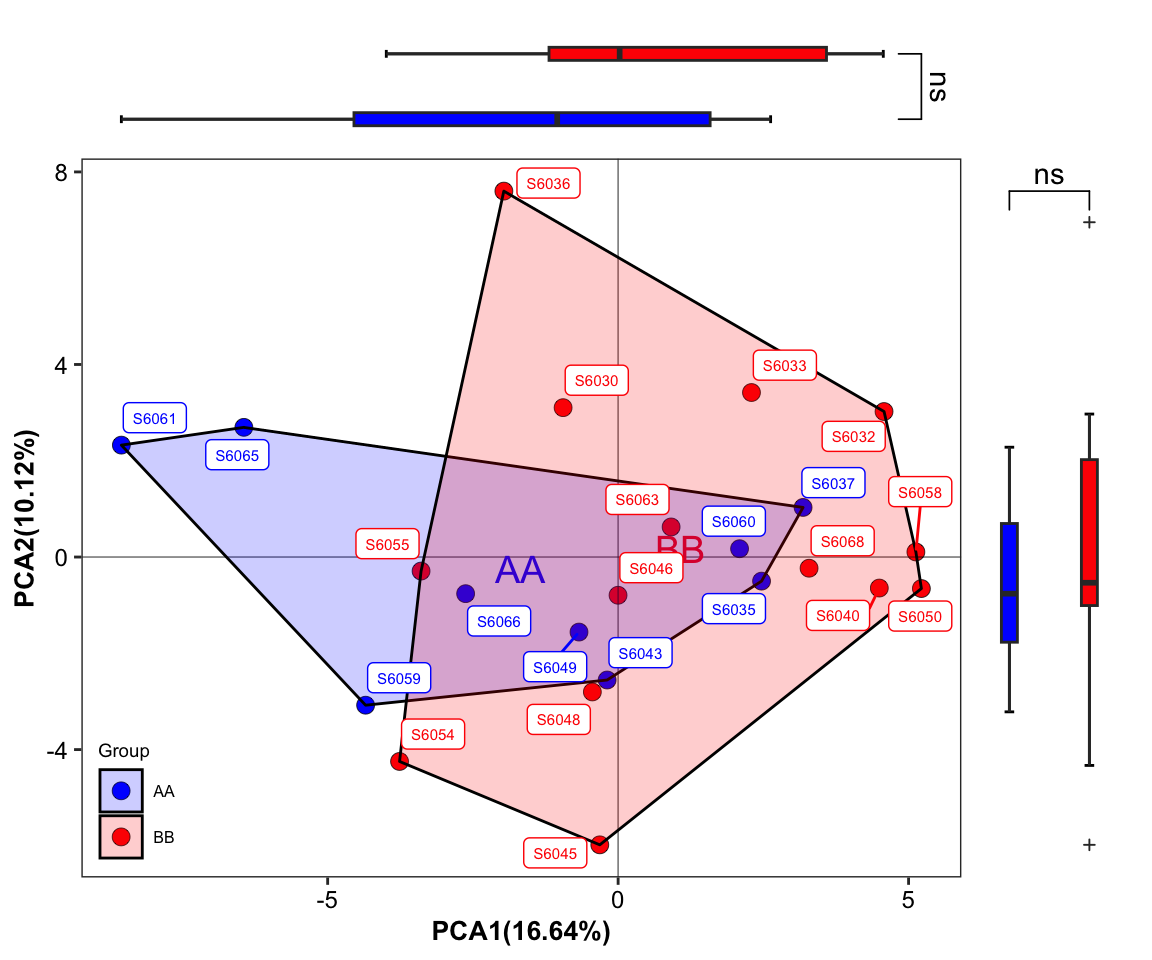
Figure 11.32: plot_Ordination (ellipse_line with SampleID sideboxplot)
- plot with SampleID and sideboxplot and setting group colors and shape
data("amplicon_ps")
amplicon_ps_genus <- summarize_taxa(ps = amplicon_ps,
taxa_level = "Genus")
amplicon_res_ordination <- run_ordination(
ps = amplicon_ps_genus,
group = "SampleType",
method = "PCoA")## [1] "Pvalue of beta dispersion less than 0.05"plot_Ordination(ResultList = amplicon_res_ordination,
group = "SampleType",
shape_column = "ReportedAntibioticUsage",
shape_values = c(16, 17),
circle_type = "ellipse_line",
sidelinechart = FALSE,
sideboxplot = TRUE,
sample = TRUE)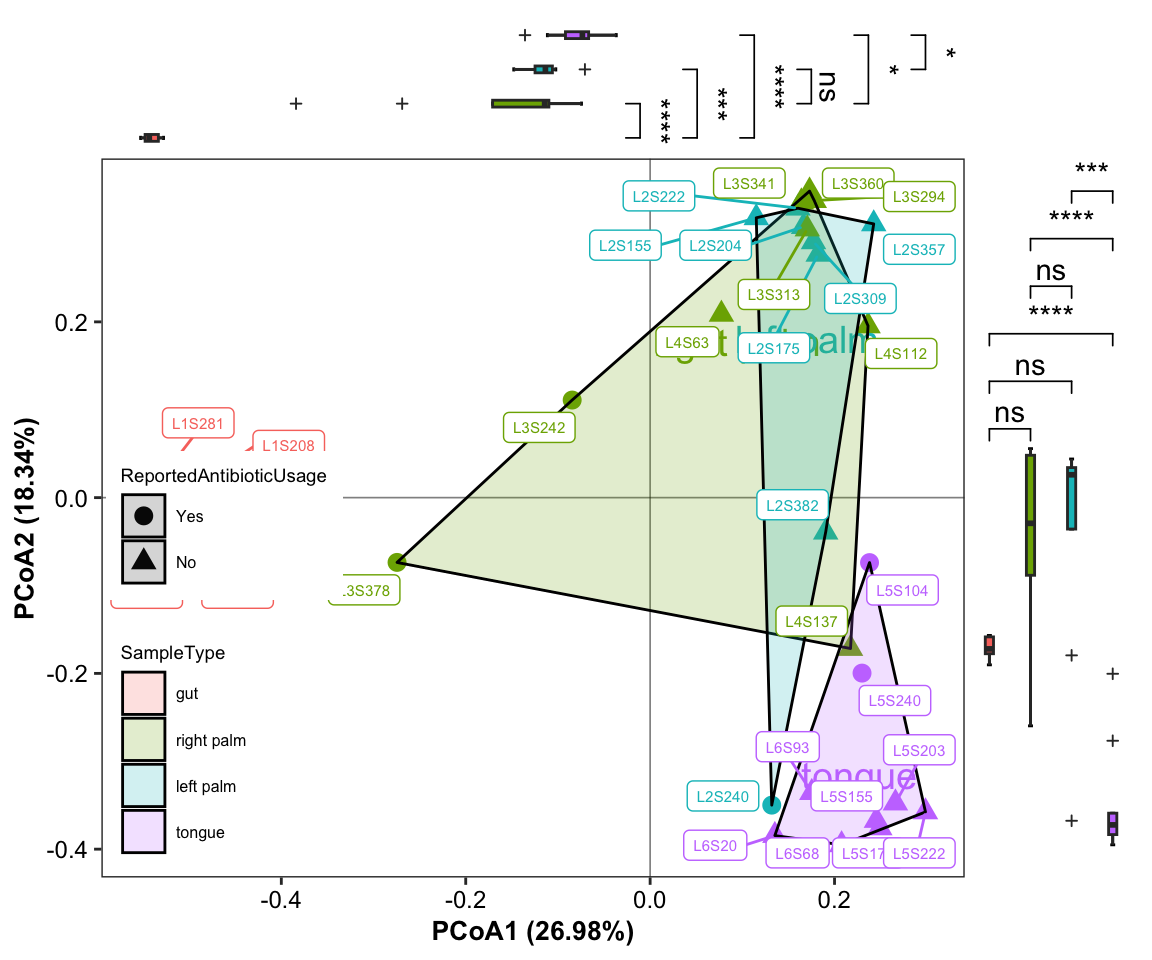
Figure 11.33: plot_Ordination (ellipse_line with SampleID sideboxplot)
11.9 plot_ggbiplot
- biplot with topN dominant taxa
plot_ggbiplot(ResultList = ordination_PCA,
group = "Group",
group_color = c("blue", "red"),
topN = 5,
ellipse = TRUE,
labels = "SampleID")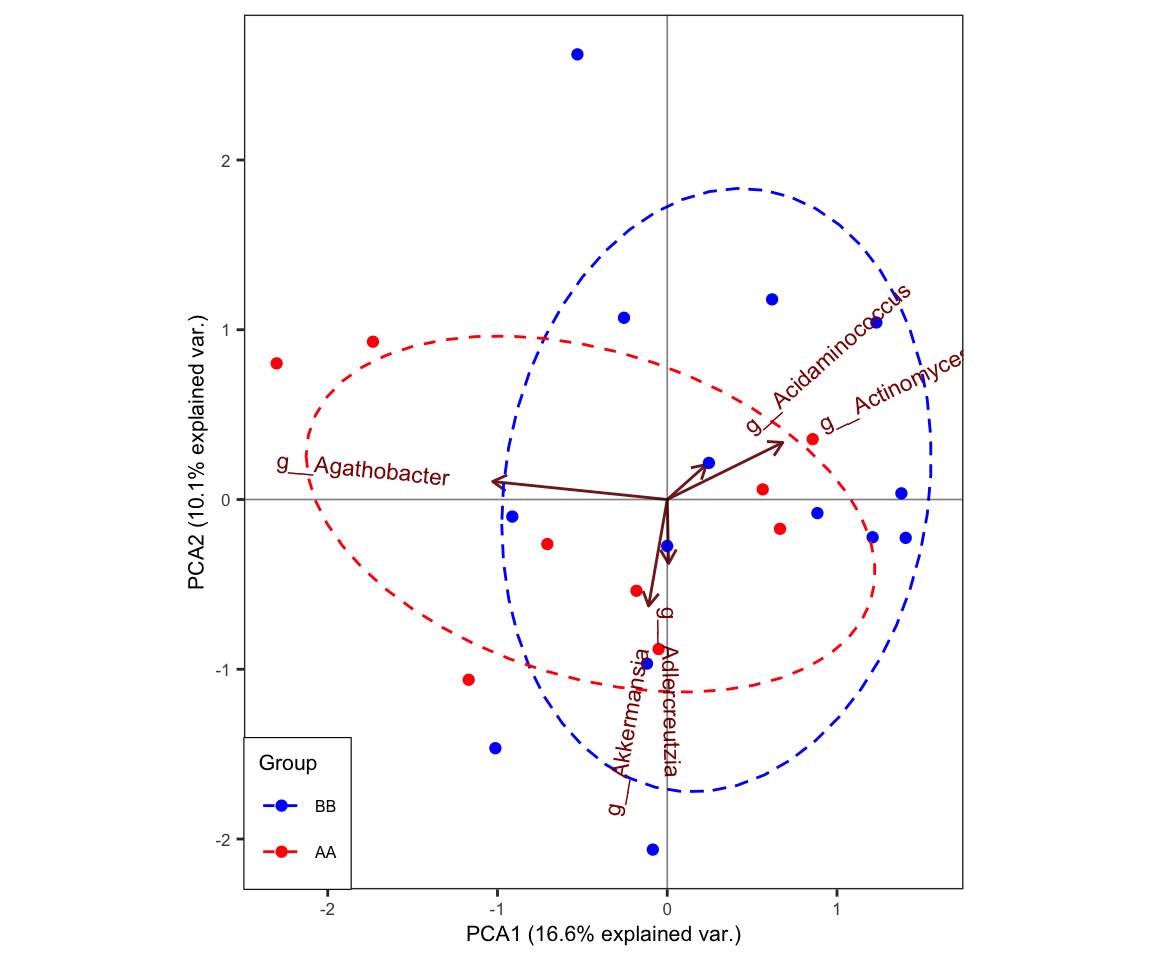
Figure 11.34: plot_ggbiplot (biplot)
11.10 plot_corrplot
dada2_beta <- run_beta_diversity(ps = dada2_ps_rarefy,
method = "bray")
plot_distance_corrplot(datMatrix = dada2_beta$BetaDistance)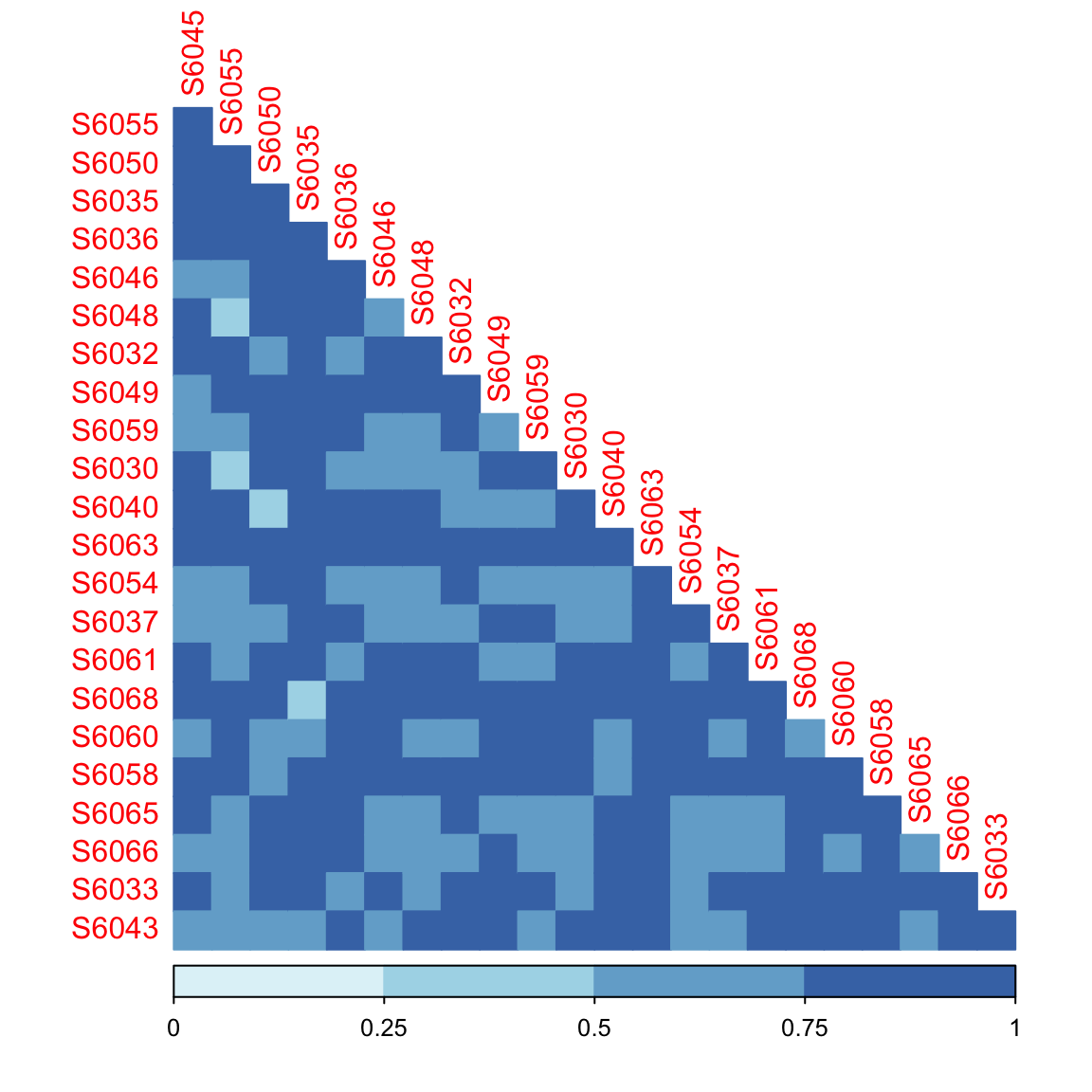
Figure 11.35: plot_corrplot (distance)
11.11 plot_2DA_venn
da_wilcox <- run_wilcox(
ps = dada2_ps_genus_filter_trim,
group = "Group",
group_names = c("AA", "BB"))
da_ttest <- run_ttest(
ps = dada2_ps_genus_filter_trim,
group = "Group",
group_names = c("AA", "BB"))
DA_venn_res <- plot_2DA_venn(
daTest1 = da_wilcox,
daTest2 = da_ttest,
datType1 = "AA vs BB(wilcox)",
datType2 = "AA vs BB(t-test)",
group_names = c("AA", "BB"),
Pvalue_name = "Pvalue",
logFc_name1 = "Log2FoldChange (Rank)\nAA_vs_BB",
logFc_name2 = "Log2FoldChange (Mean)\nAA_vs_BB",
Pvalue_cutoff = 0.8,
logFC_cutoff = 0.2)
DA_venn_res$pl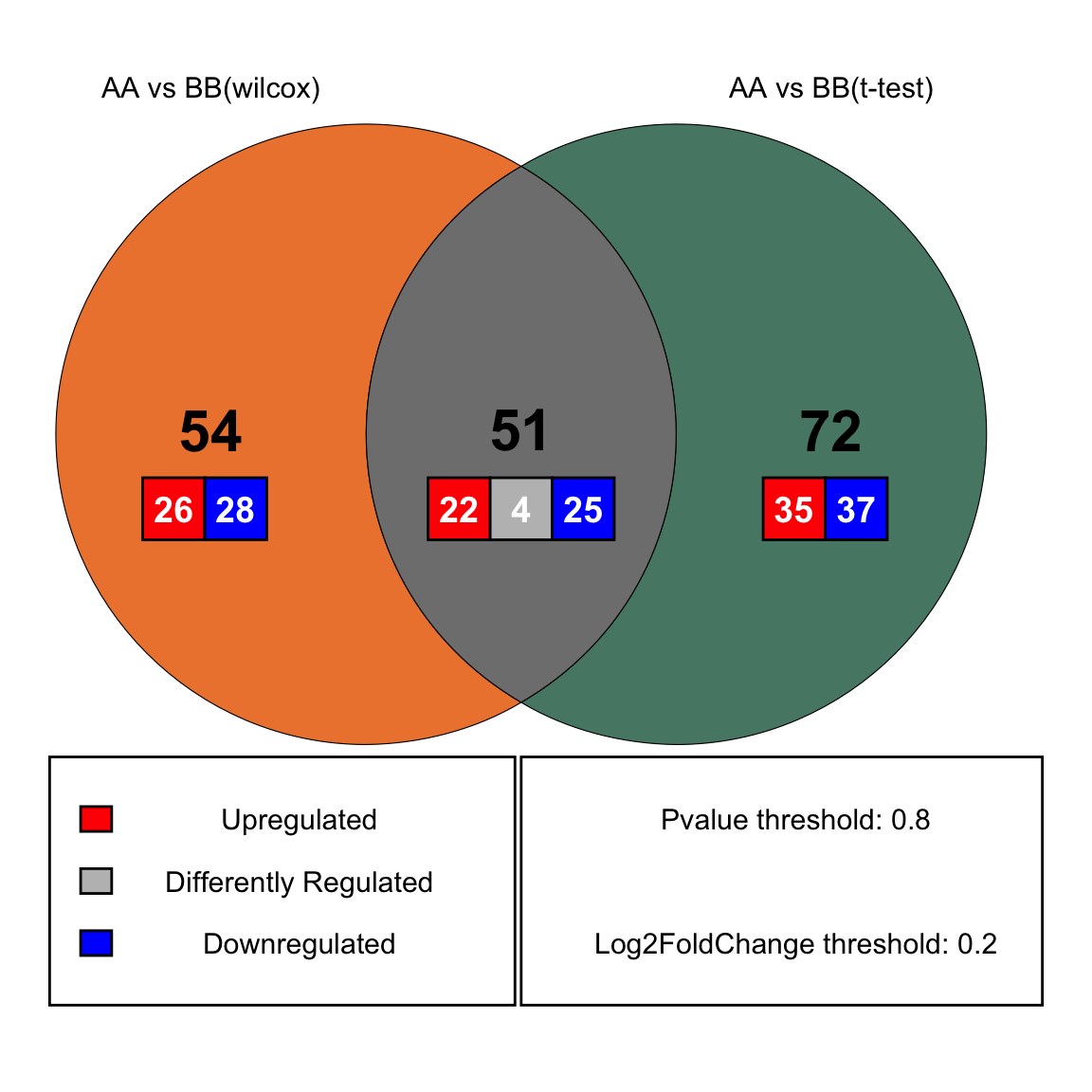
Figure 11.36: plot_2DA_venn (wilcox vs t_test)
11.12 plot the DA results from the significant taxa by double barplot
data("amplicon_ps")
DA_res <- run_wilcox(
ps = amplicon_ps,
taxa_level = "Family",
group = "SampleType",
group_names = c("tongue", "gut"))
plot_double_barplot(data = DA_res,
x_index = "Log2FoldChange (Rank)\ntongue_vs_gut",
x_index_cutoff = 1,
y_index = "AdjustedPvalue",
y_index_cutoff = 0.05)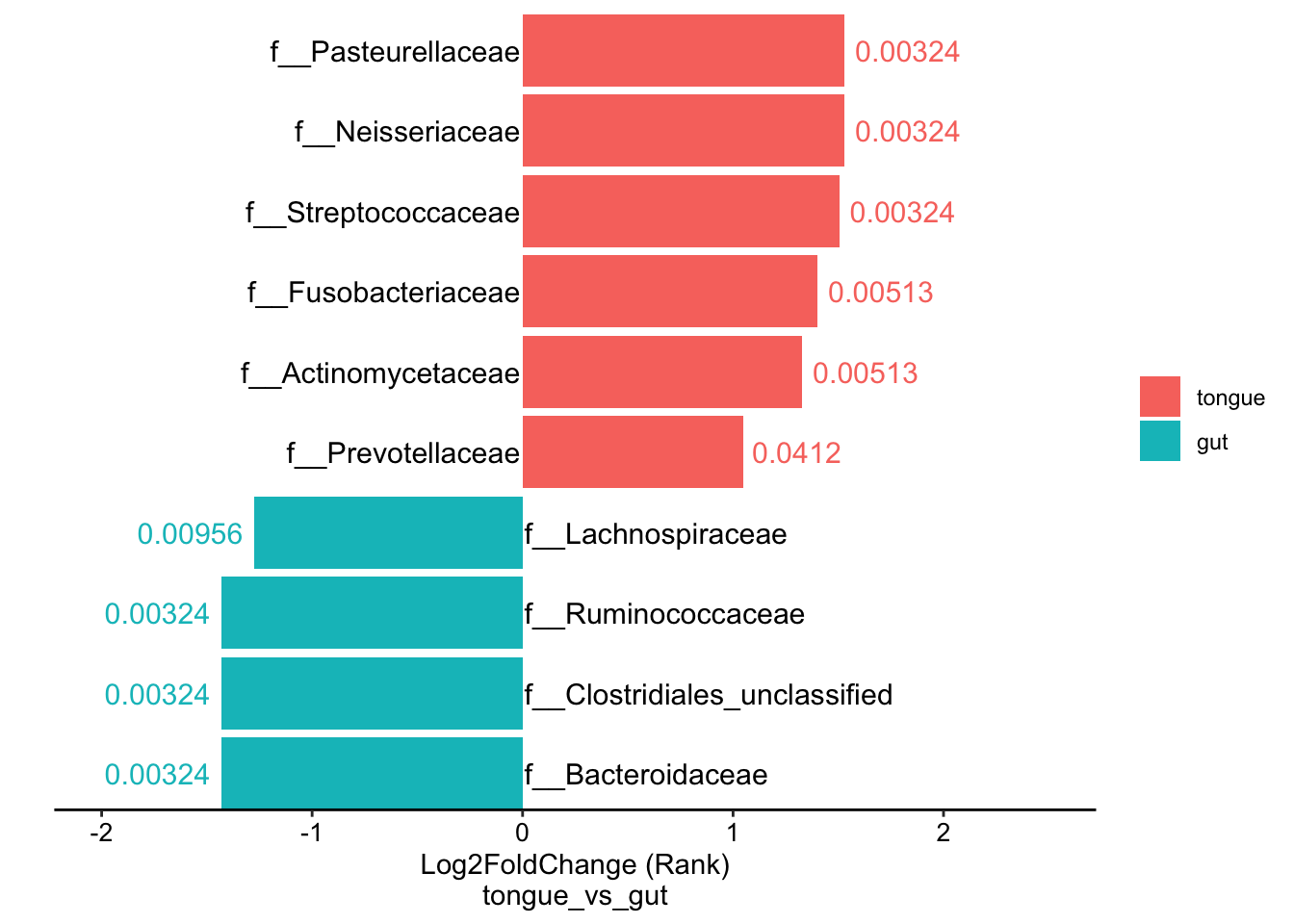
Figure 11.37: double barplot for DA results
11.13 plot_stacked_bar_XIVZ
- Minimum usage: plot in relative abundance
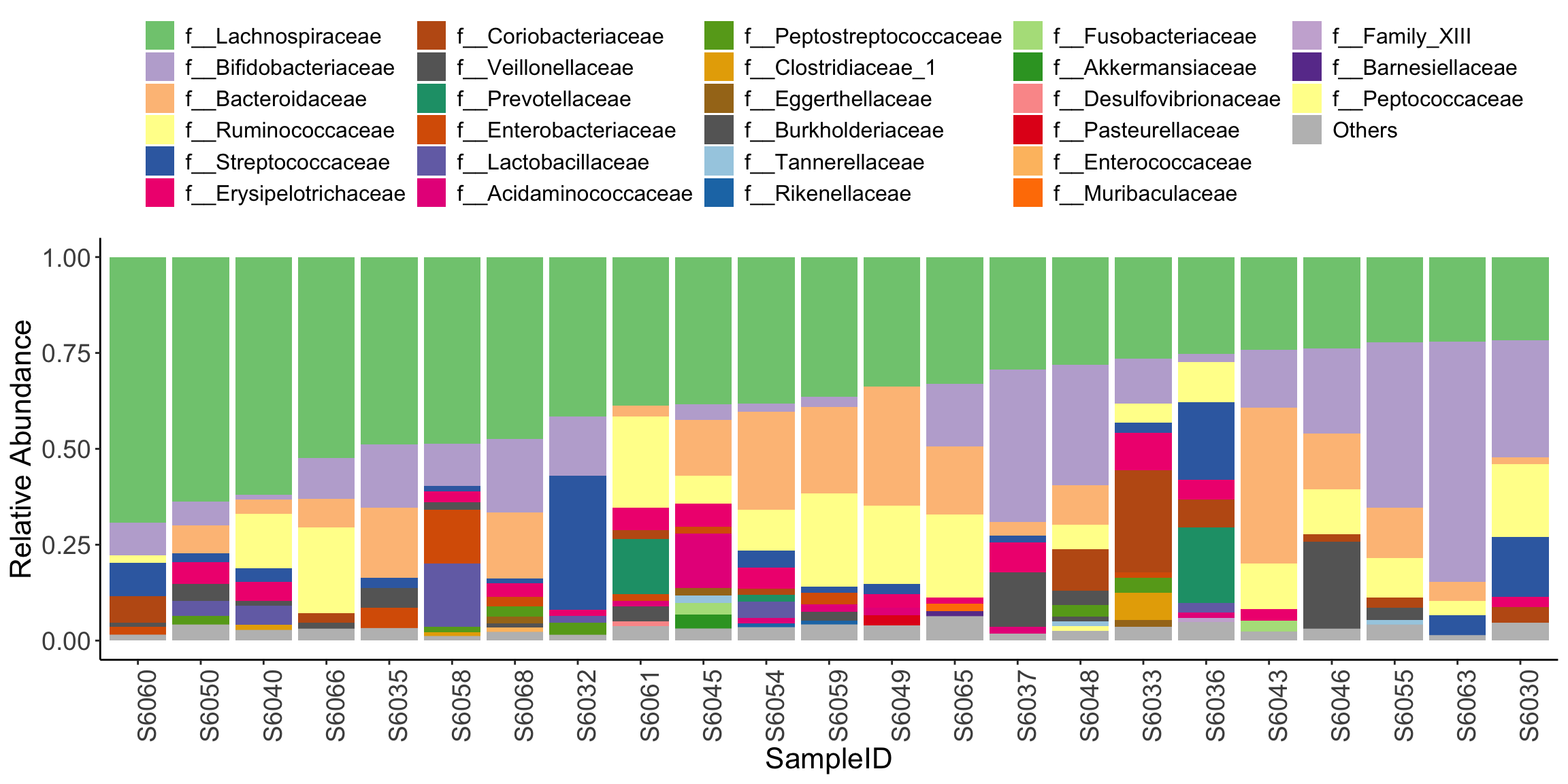
Figure 11.38: plot_stacked_bar_XIVZ (test1)
- Set feature parameter to show feature information
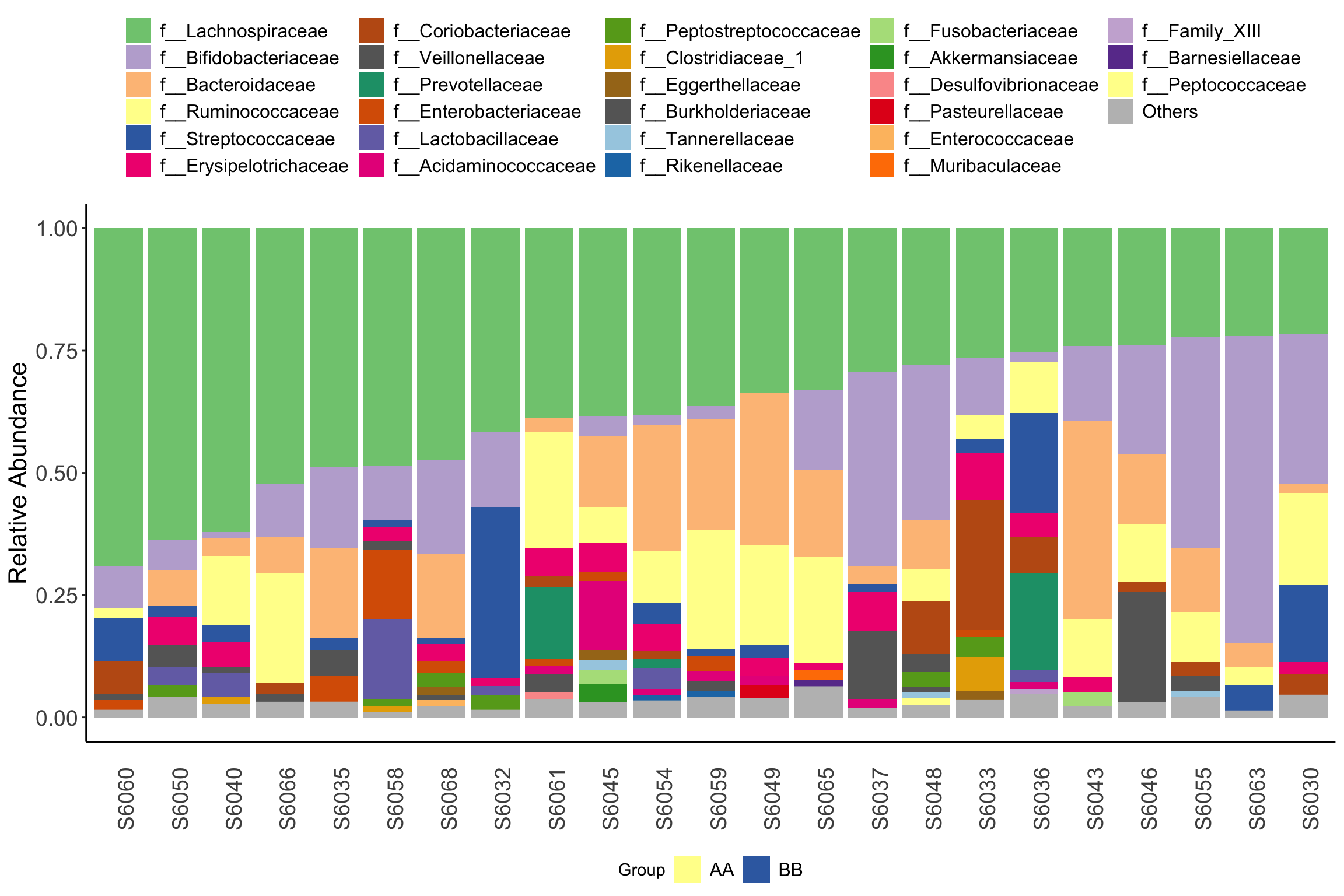
Figure 11.39: plot_stacked_bar_XIVZ (test2)
- Pass ordered sample names to order parameter to plot in specific order
metadata <- phyloseq::sample_data(dada2_ps_rarefy) %>%
data.frame() %>%
dplyr::arrange(Group)
plot_stacked_bar_XIVZ(phyloseq = dada2_ps_rarefy,
level = "Family",
feature = "Group",
order = rownames(metadata))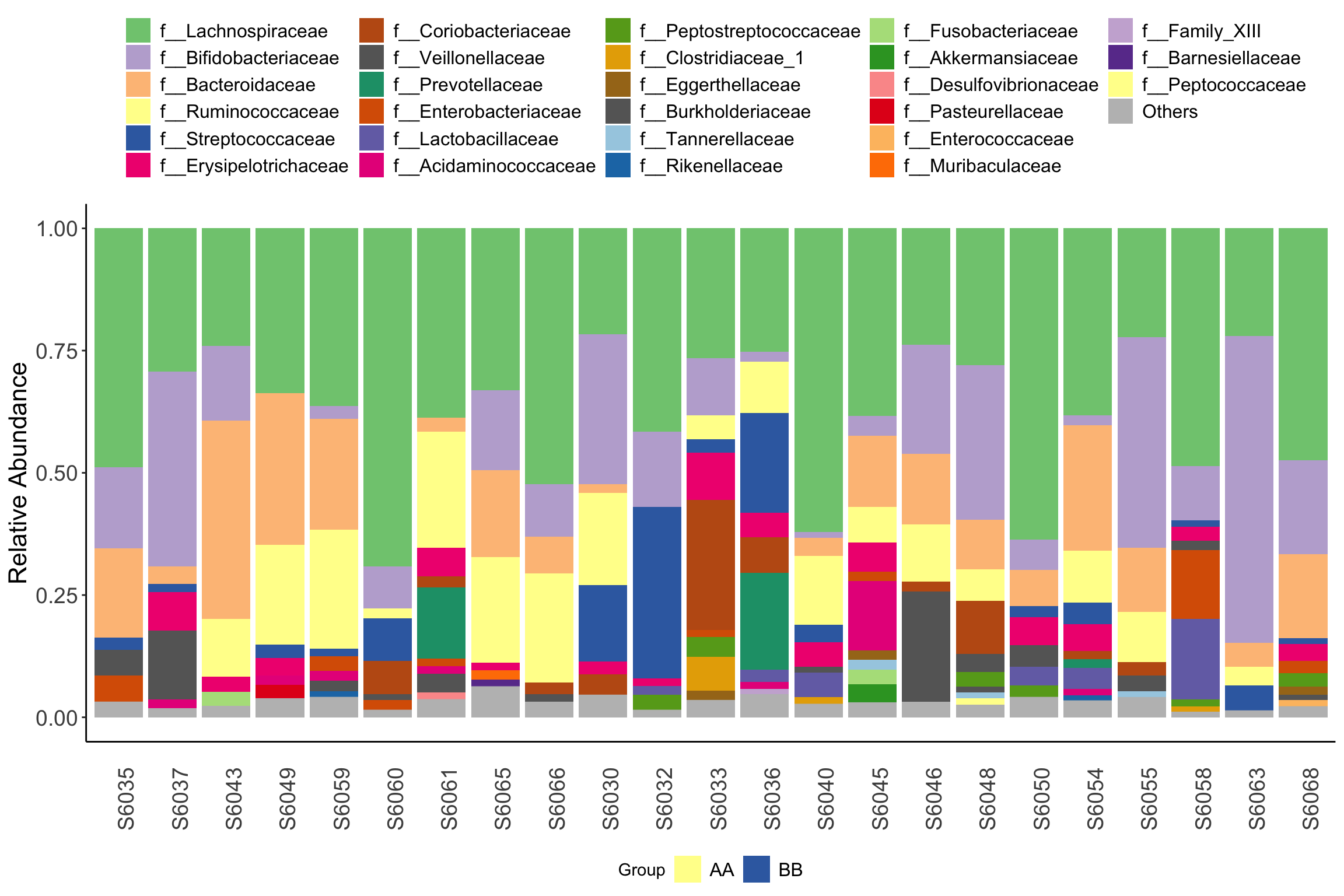
Figure 11.40: plot_stacked_bar_XIVZ (test3)
- Use facet_wrap(vars(), scale=“free”) funciton to facet stacked barplot
plot_stacked_bar_XIVZ(phyloseq = dada2_ps_rarefy,
level = "Family",
relative_abundance = TRUE,
order = rownames(metadata)) +
facet_wrap(vars(Group), scale="free")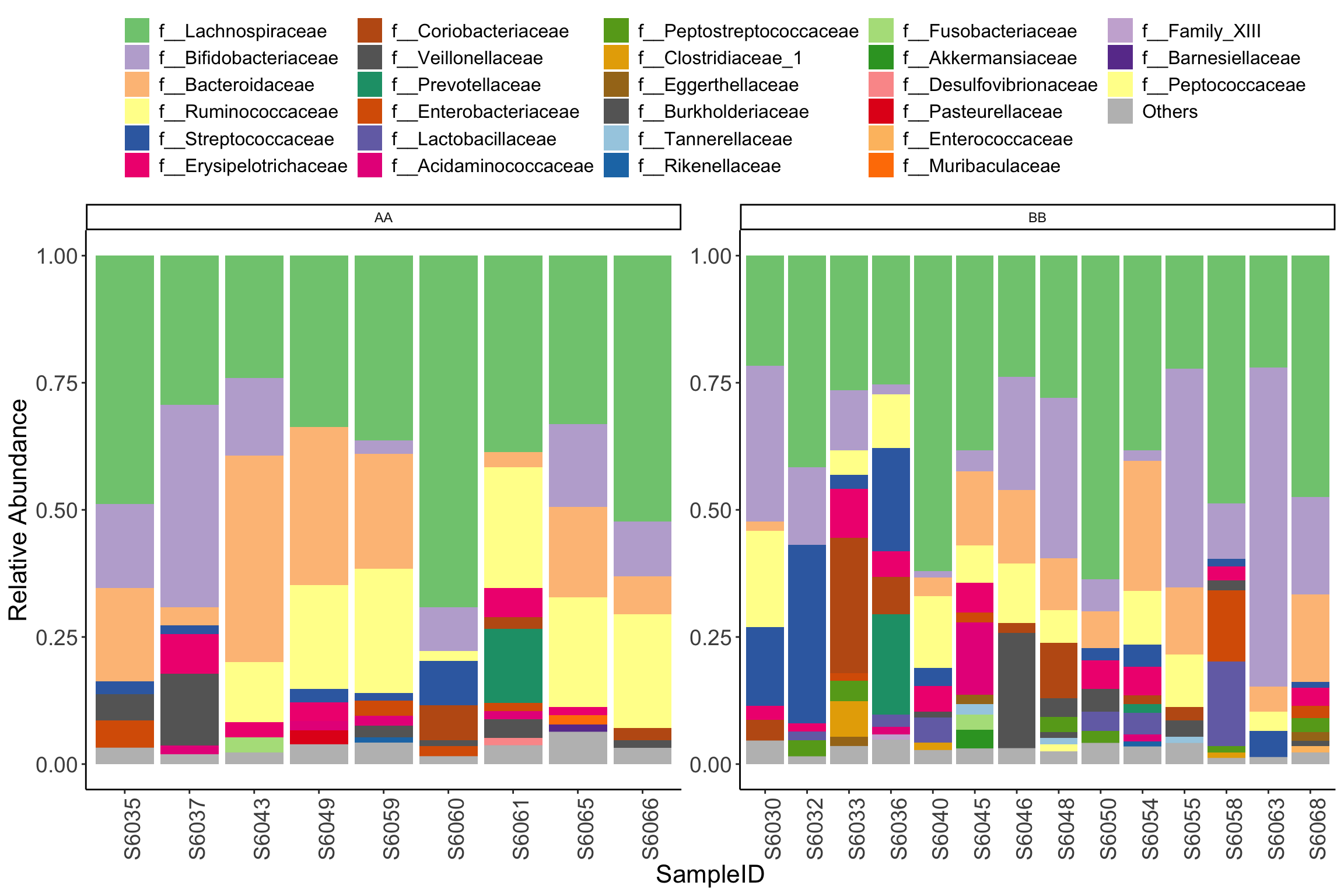
Figure 11.41: plot_stacked_bar_XIVZ (test4)
11.14 plot_StackBarPlot
plot_StackBarPlot provides too many parameters for users to display the Stacked barplot of microbial composition by using ggplot2 format. Here is the ordinary pattern. More details to see help(plot_StackBarPlot).
- Ordinary pattern
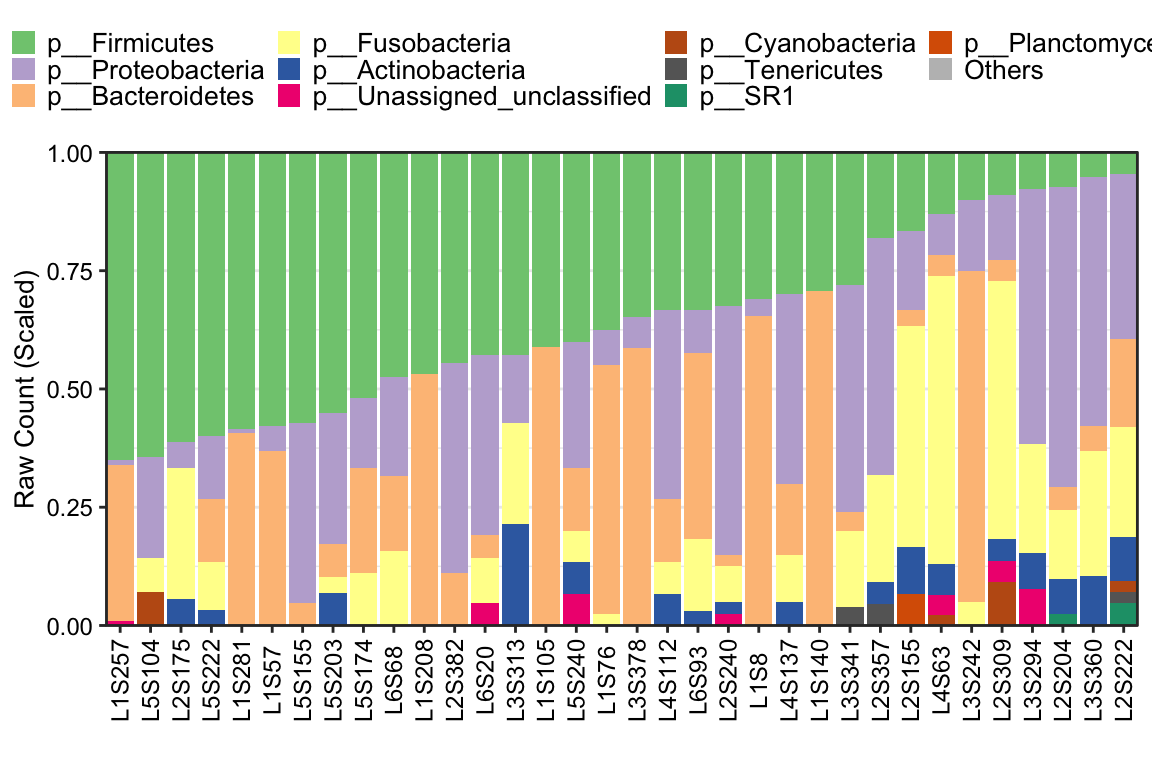
Figure 11.42: plot_StackBarPlot(Ordinary pattern)
- Metadata with
SampleTypephenotype
## [1] "This palatte have 20 colors!"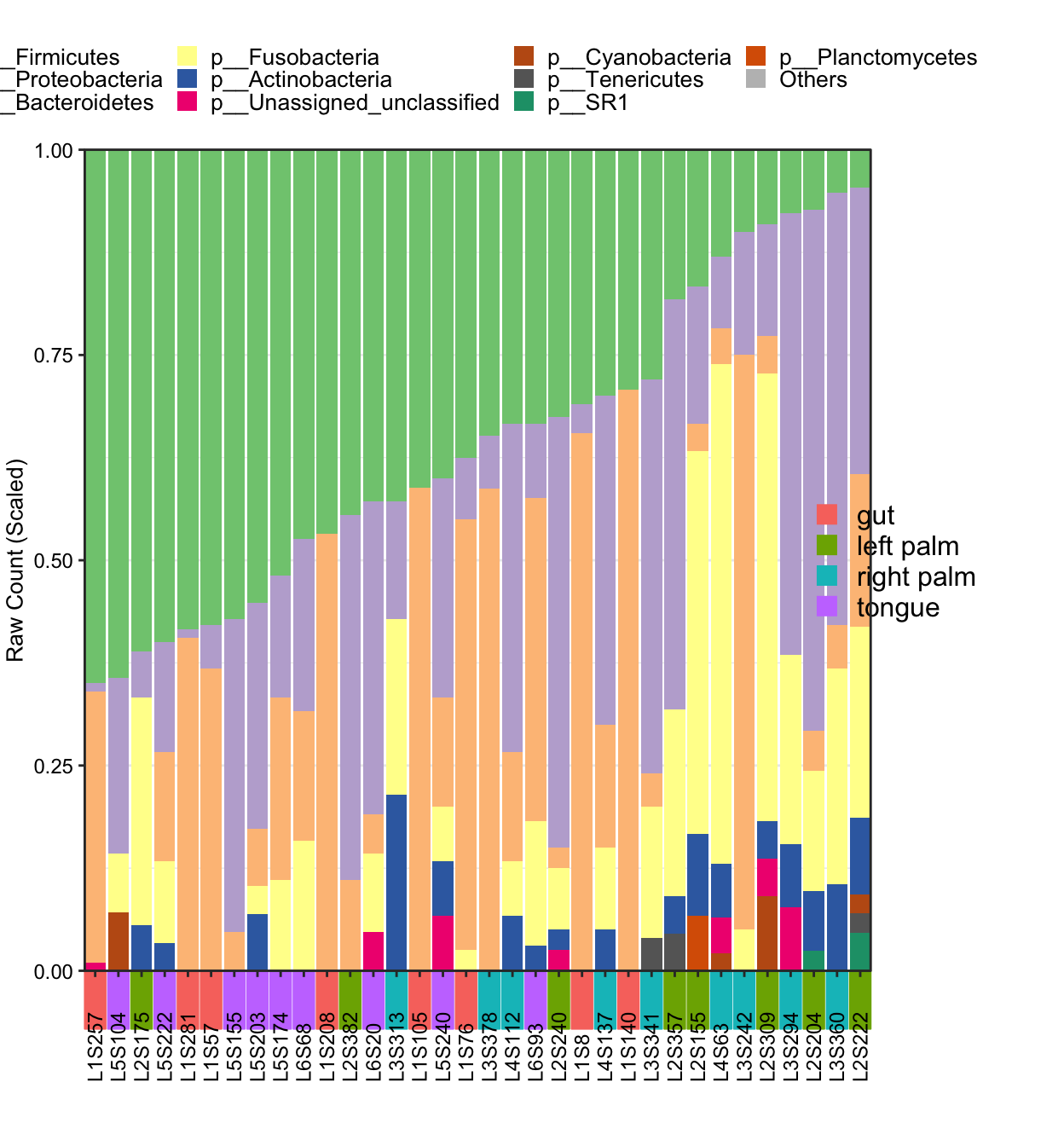
Figure 11.43: plot_StackBarPlot (Metadata with group)
- Metadata with
SampleTypephenotype in cluster mode
plot_StackBarPlot(
ps = amplicon_ps_rarefy,
taxa_level = "Phylum",
group = "SampleType",
cluster = TRUE)## [1] "This palatte have 20 colors!"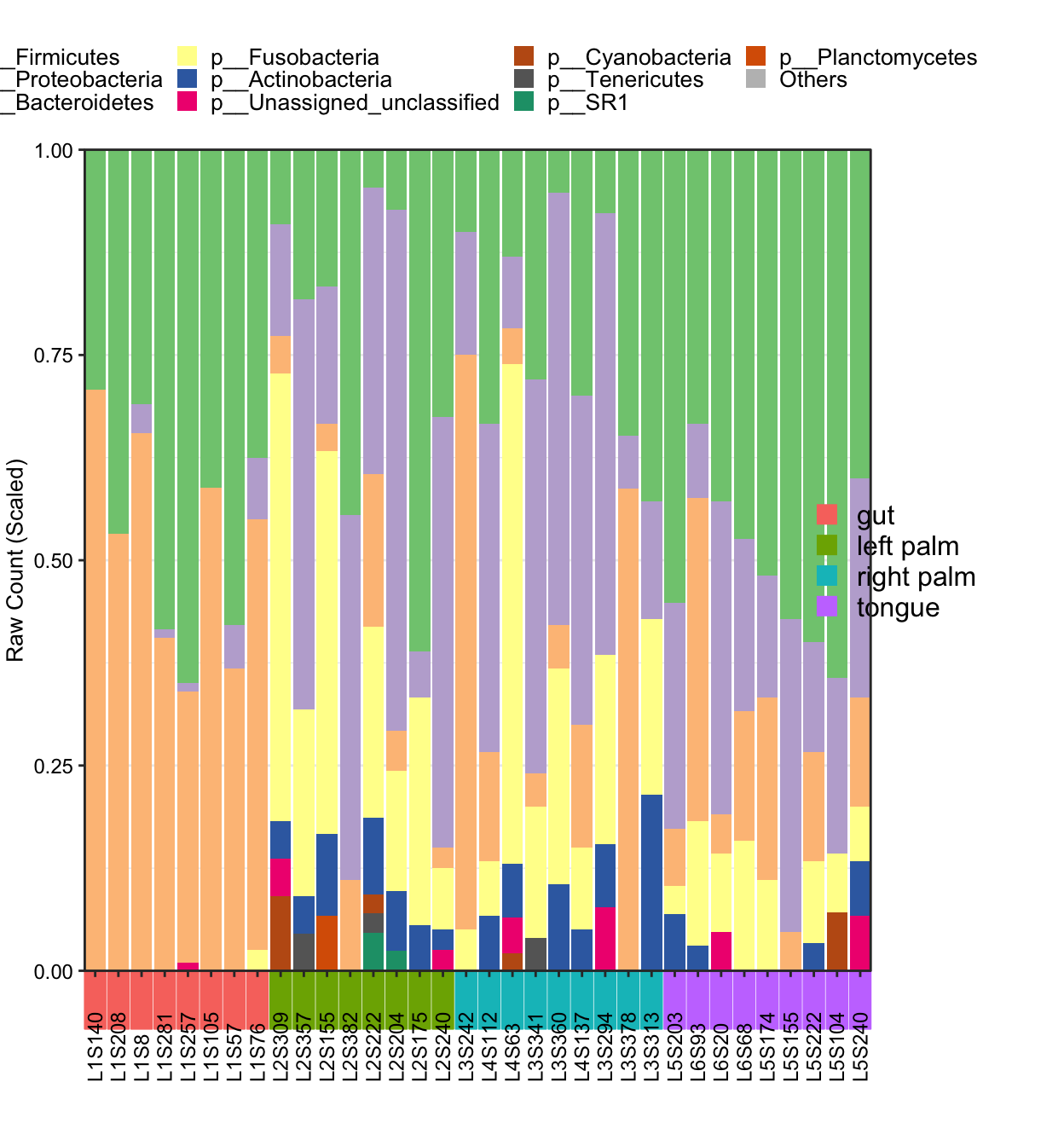
Figure 11.44: plot_StackBarPlot (Metadata with group in cluster mode)
- Metadata with
SampleTypephenotype in facet
plot_StackBarPlot(
ps = amplicon_ps_rarefy,
taxa_level = "Phylum",
group = "SampleType",
facet = TRUE)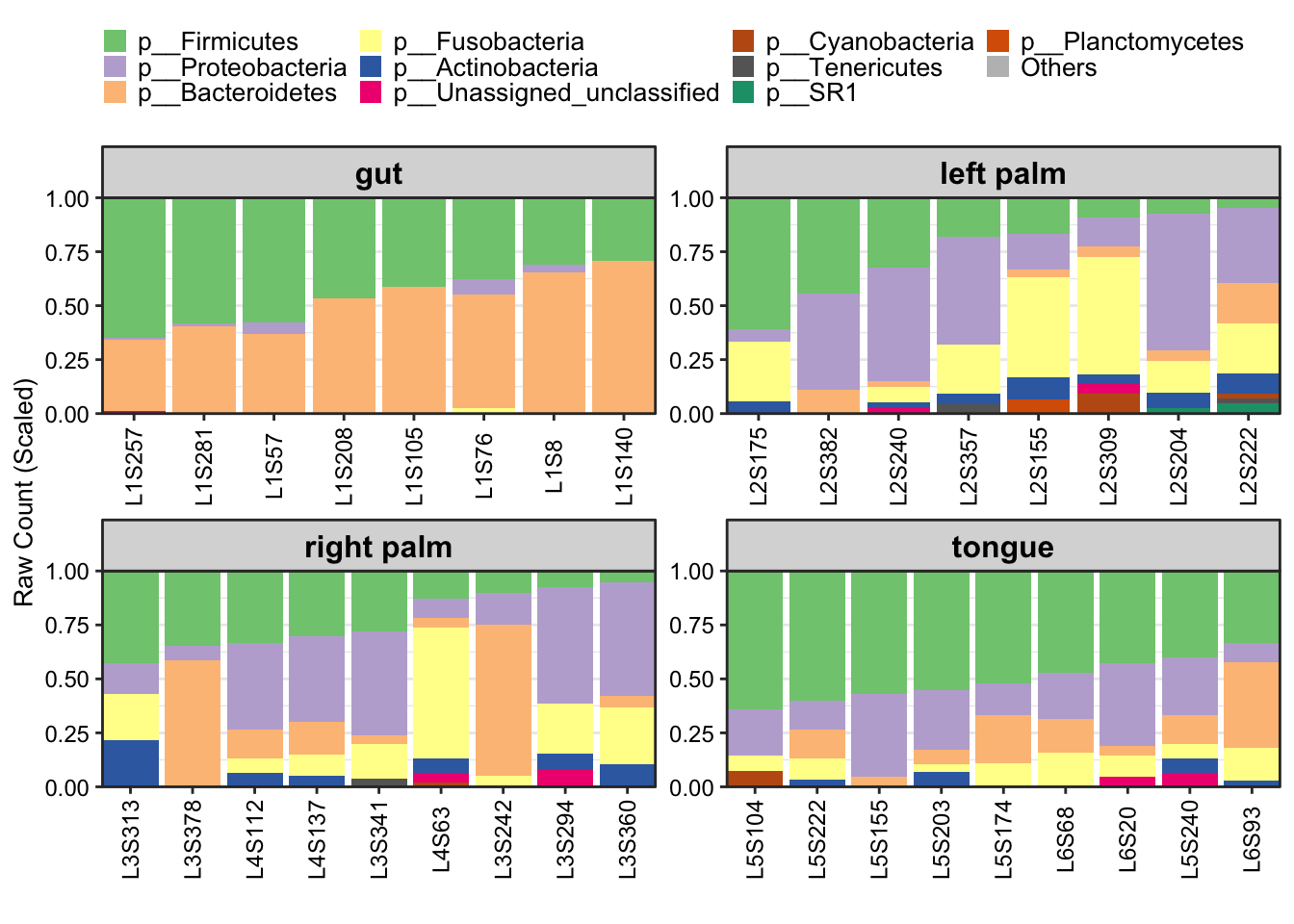
Figure 11.45: plot_StackBarPlot (Metadata with group in facet)
- Metadata with two groups to display samples
plot_StackBarPlot(
ps = amplicon_ps_rarefy,
taxa_level = "Order",
group = "SampleType",
subgroup = "Year")## [1] "This palatte have 19 colors!"
## [1] "This palatte have 20 colors!"
## [1] "This palatte have 20 colors!"
## [1] "This palatte have 20 colors!"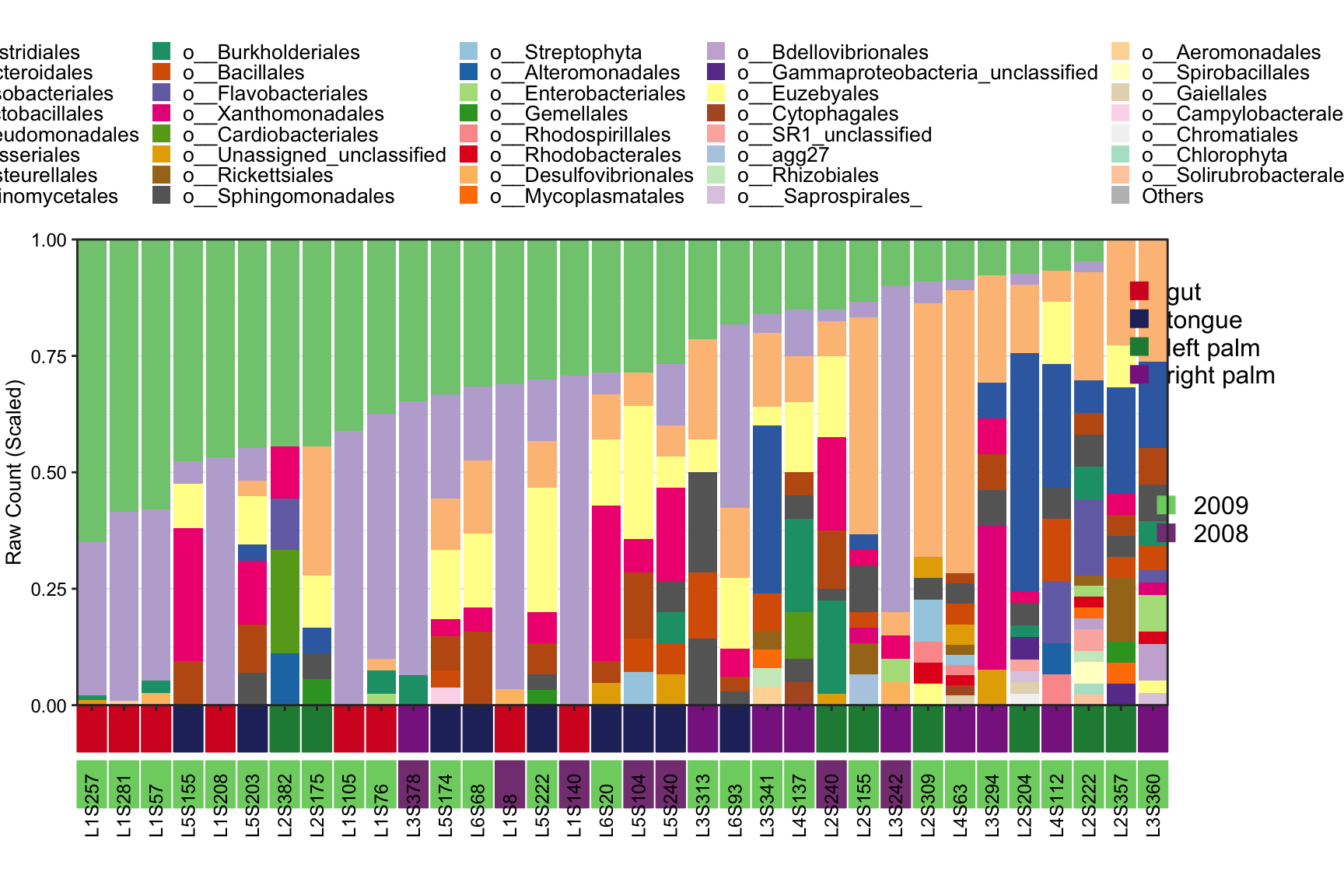
Figure 11.46: plot_StackBarPlot (Metadata with two groups ot display samples)
- Metadata with three groups to display samples
plot_StackBarPlot(
ps = amplicon_ps_rarefy,
taxa_level = "Order",
group = "SampleType",
subgroup = c("Year", "Month"))## [1] "This palatte have 19 colors!"
## [1] "This palatte have 20 colors!"
## [1] "This palatte have 20 colors!"
## [1] "This palatte have 20 colors!"
## [1] "This palatte have 20 colors!"
## [1] "This palatte have 20 colors!"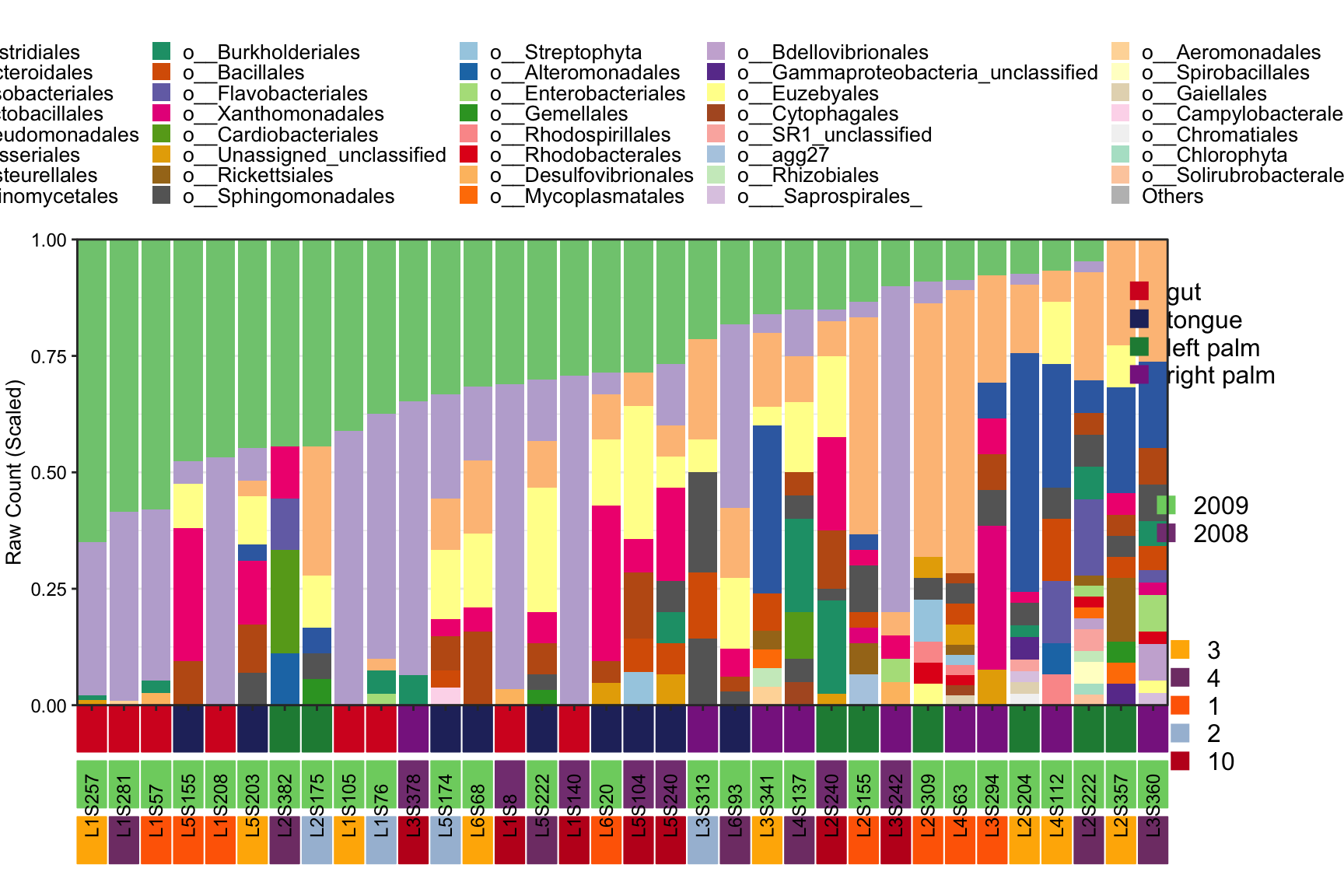
Figure 11.47: plot_StackBarPlot (Metadata with three groups ot display samples)
- Order SampleID by orderSample parameter
plot_StackBarPlot(
ps = amplicon_ps_rarefy,
taxa_level = "Order",
group = "SampleType",
orderSample = phyloseq::sample_names(amplicon_ps)[1:10])## [1] "This palatte have 20 colors!"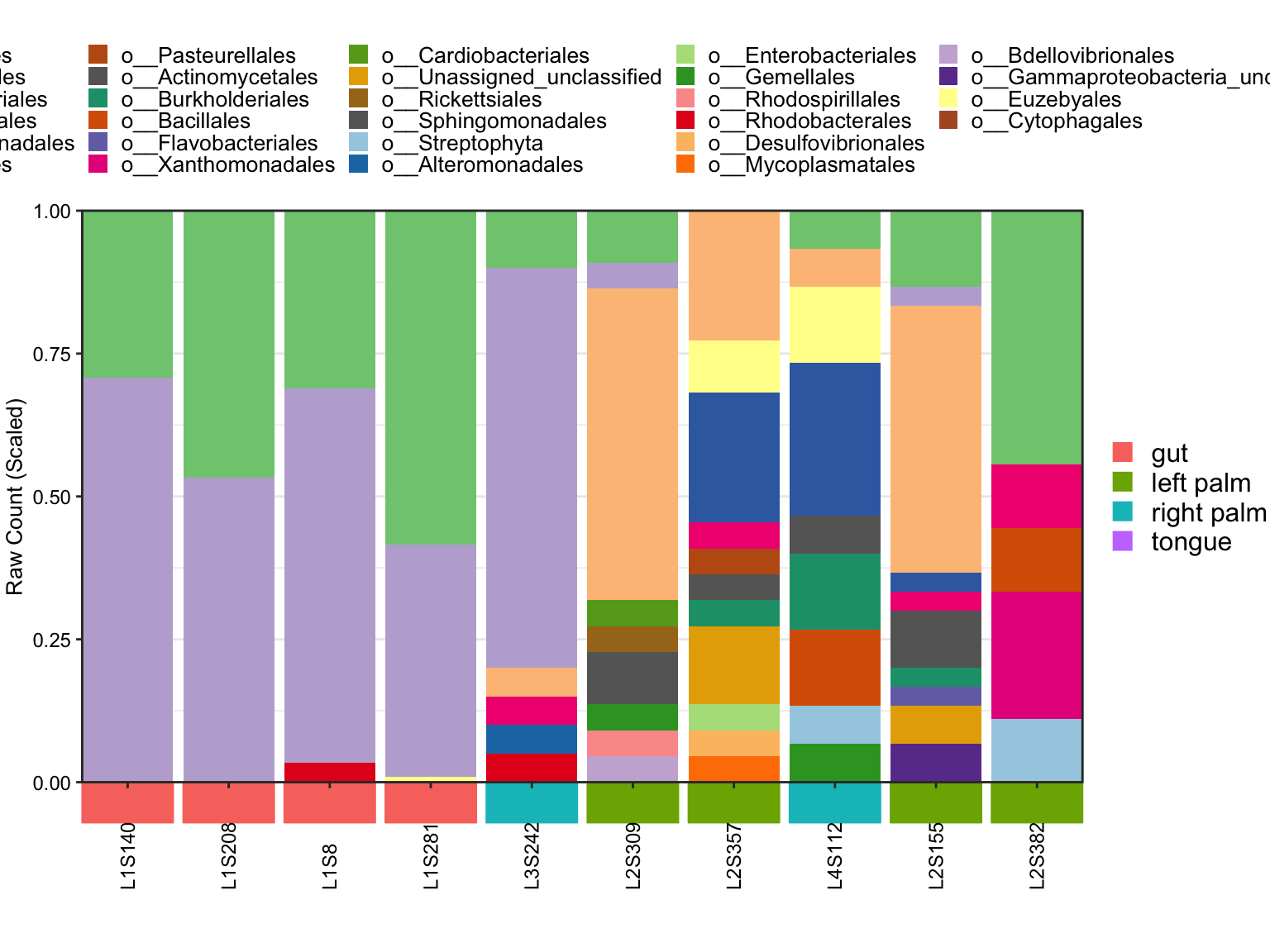
Figure 11.48: Stacked barplot with Ordered Samples
- Hide sample names by sample_label parameter
plot_StackBarPlot(
ps = amplicon_ps_rarefy,
taxa_level = "Order",
group = "SampleType",
orderSample = phyloseq::sample_names(amplicon_ps)[1:10],
sample_label = FALSE)## [1] "This palatte have 20 colors!"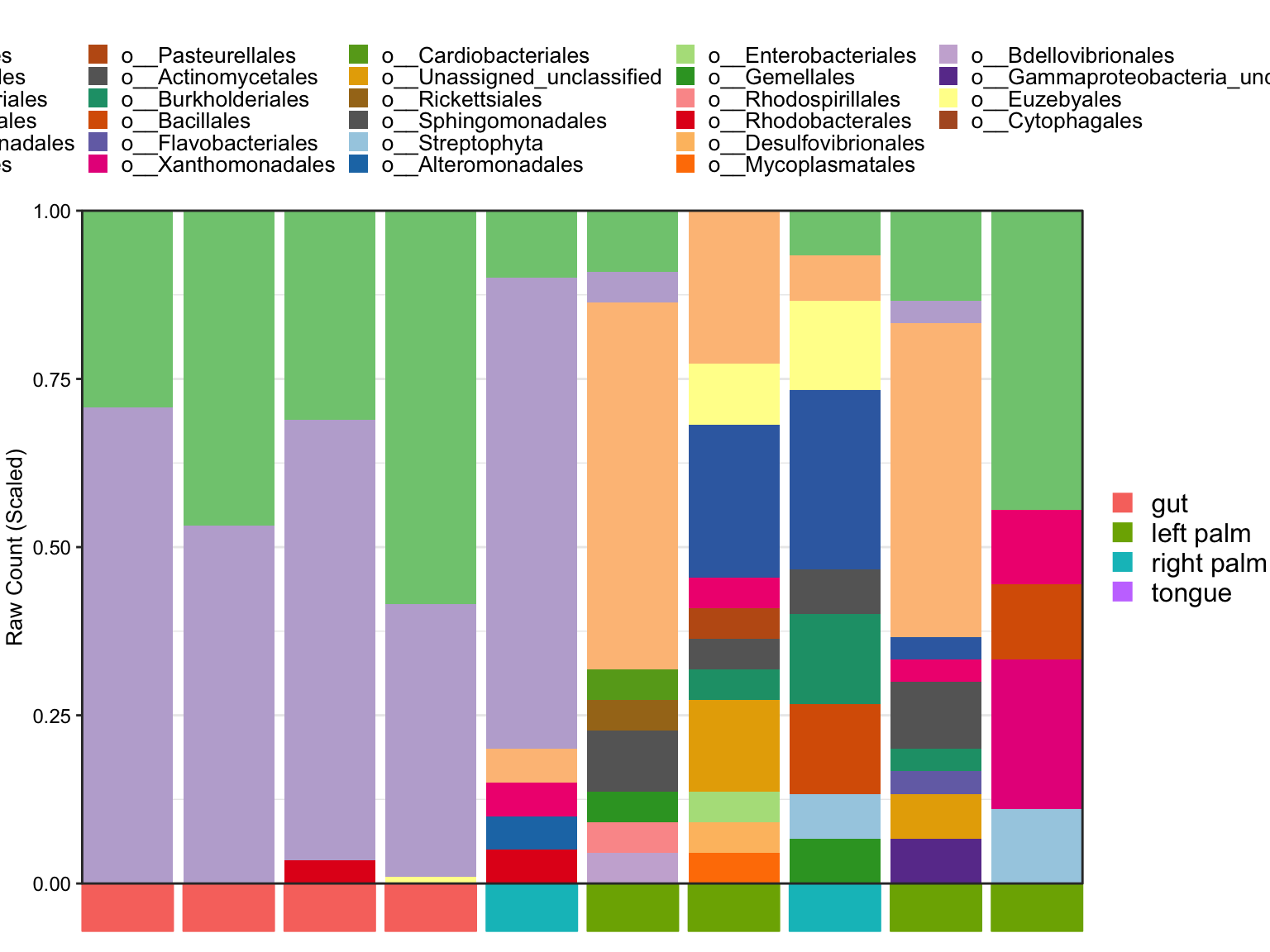
Figure 11.49: Stacked barplot with hiding Samples’ names
11.15 Color Palettes
11.15.1 Wes Anderson Palettes
Wes Anderson Palettes is from wesanderson package and we have integrated it into XMAS2.0.
data("amplicon_ps")
dat_alpha <- run_alpha_diversity(ps = amplicon_ps, measures = c("Shannon", "Chao1"))
# origin
pl_origin <- plot_boxplot(data = dat_alpha,
y_index = "Shannon",
group = "SampleType",
do_test = TRUE,
cmp_list = list(c("gut", "right palm"), c("gut", "left palm")),
method = "wilcox.test")
# Wes Anderson Palettes
pal <- wes_palette(name = "GrandBudapest1", 4, type = "discrete")
pl_wes <- plot_boxplot(data = dat_alpha,
y_index = "Shannon",
group = "SampleType",
group_color = pal,
do_test = TRUE,
cmp_list = list(c("gut", "right palm"), c("gut", "left palm")),
method = "wilcox.test")
cowplot::plot_grid(pl_origin, pl_wes,
align = "hv",
labels = c("Origin", "Wes Anderson"))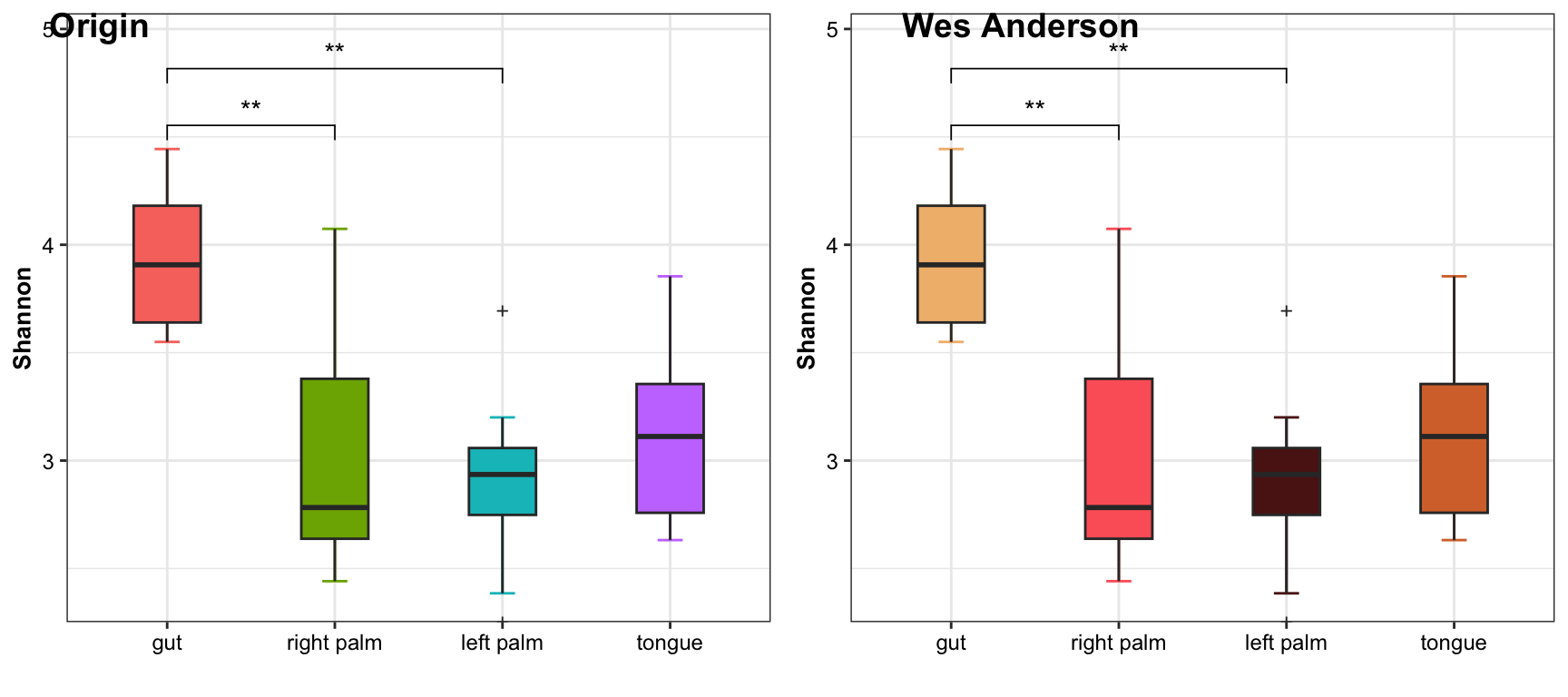
Figure 11.50: Wes Anderson Palettes
11.16 Ordination plots with ggplot2
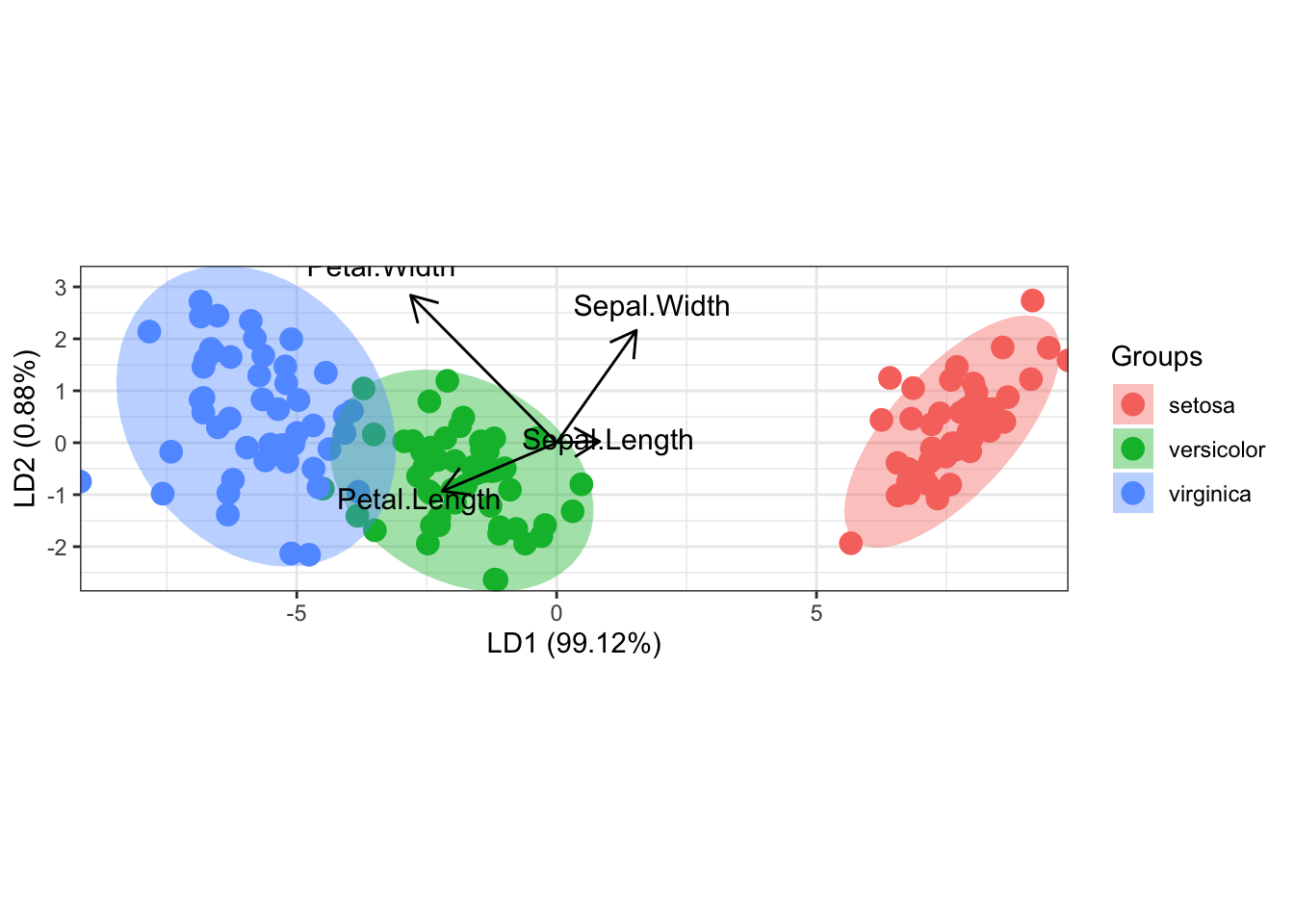
11.16.1 principal components analysis with the iris data set
ord <- prcomp(iris[, 1:4])
ggord(ord, iris$Species, cols = c('purple', 'orange', 'blue')) +
scale_shape_manual('Groups', values = c(1, 2, 3)) +
theme_classic() +
theme(legend.position = 'top')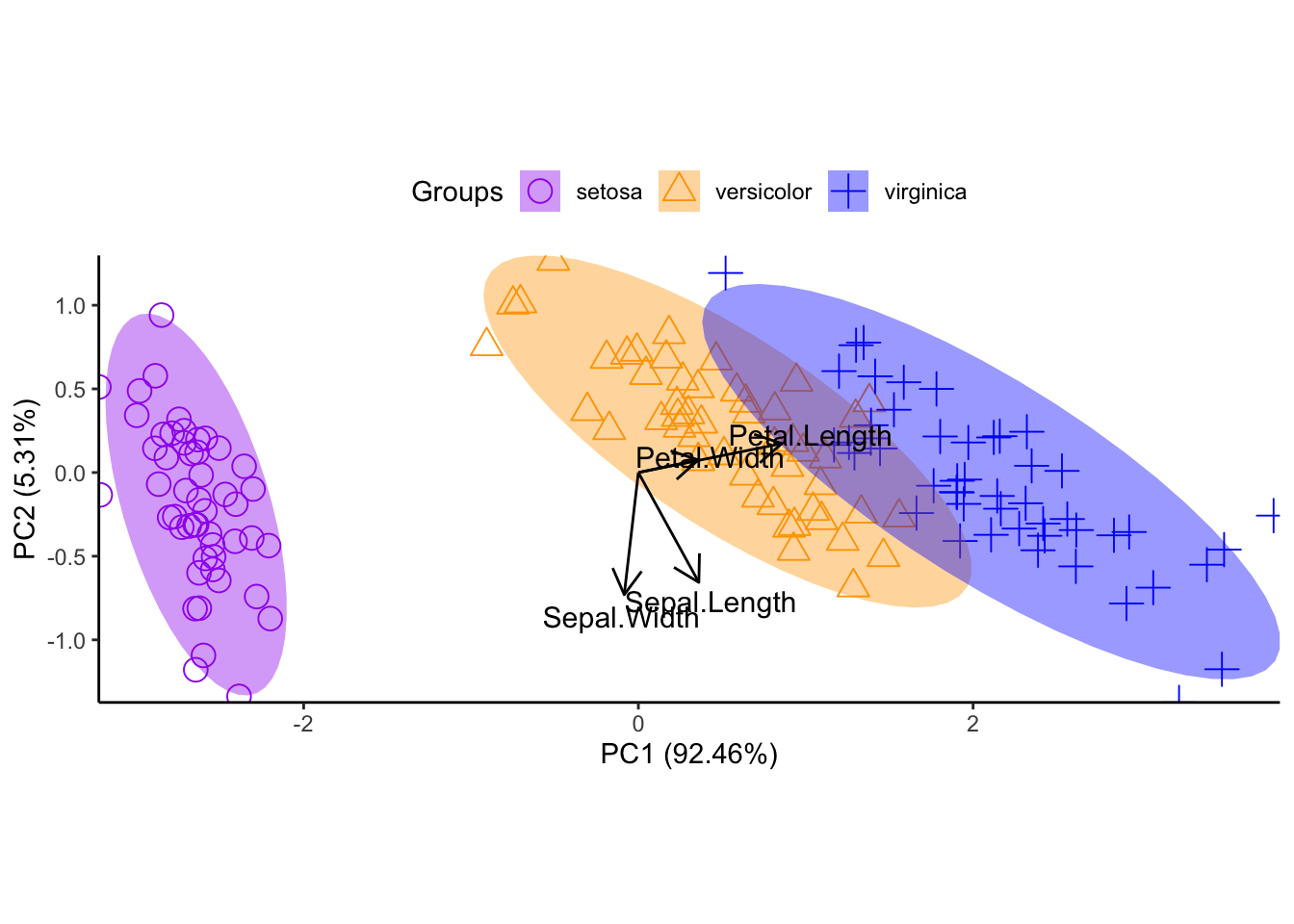
Figure 11.52: principal components analysis
11.16.2 multiple correspondence analysis with the tea dataset
data(tea, package = 'FactoMineR')
tea <- tea[, c('Tea', 'sugar', 'price', 'age_Q', 'sex')]
ord <- FactoMineR::MCA(tea[, -1], graph = FALSE)
ggord(ord, tea$Tea, parse = FALSE) # use parse = FALSE for labels with non alphanumeric characters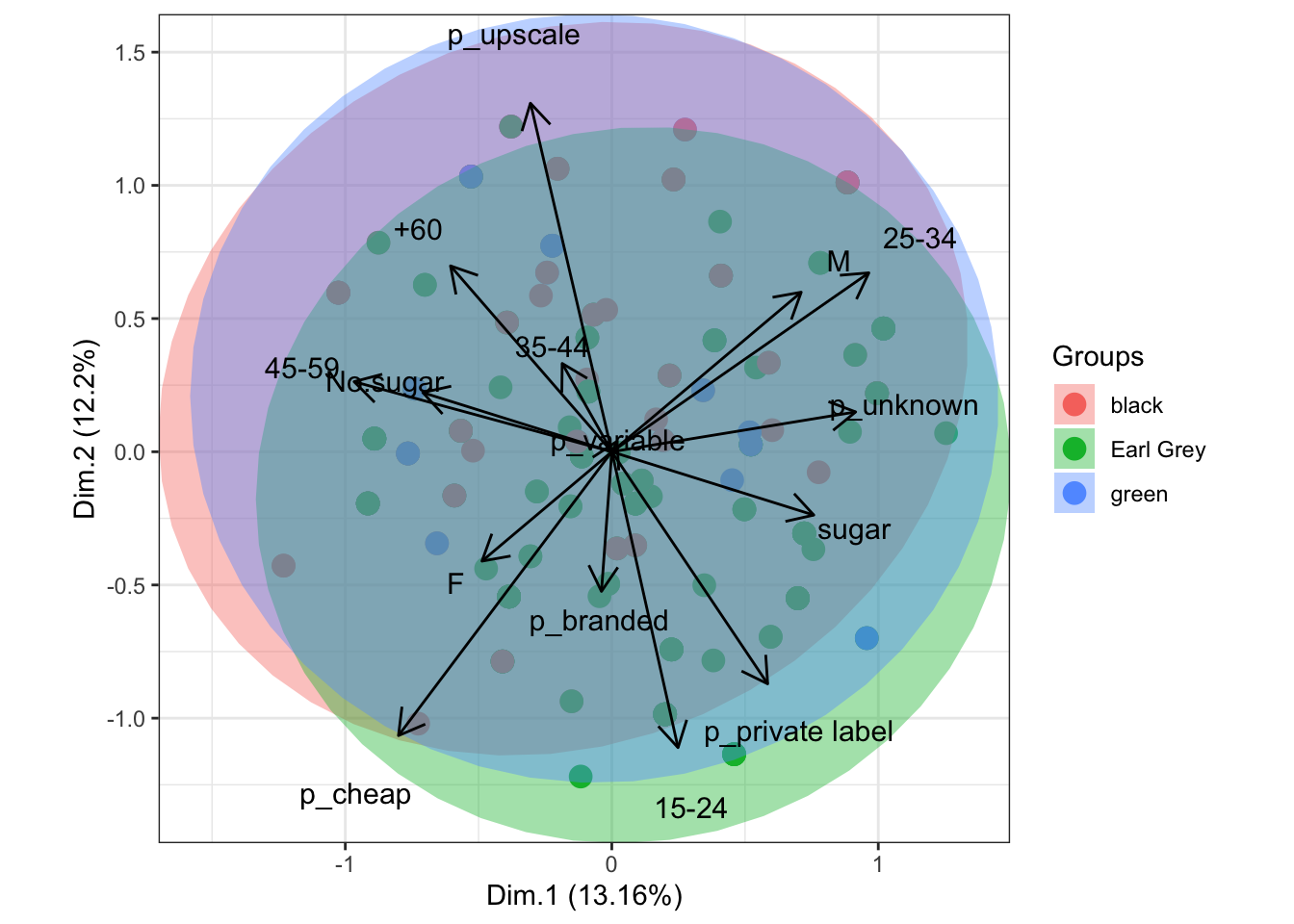
Figure 11.53: multiple correspondence analysis
11.17 Systematic Information
## ─ Session info ───────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────
## setting value
## version R version 4.1.3 (2022-03-10)
## os macOS Monterey 12.2.1
## system x86_64, darwin17.0
## ui RStudio
## language (EN)
## collate en_US.UTF-8
## ctype en_US.UTF-8
## tz Asia/Shanghai
## date 2023-11-30
## rstudio 2023.09.0+463 Desert Sunflower (desktop)
## pandoc 3.1.1 @ /Applications/RStudio.app/Contents/Resources/app/quarto/bin/tools/ (via rmarkdown)
##
## ─ Packages ───────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────
## package * version date (UTC) lib source
## abind 1.4-5 2016-07-21 [2] CRAN (R 4.1.0)
## ade4 1.7-22 2023-02-06 [2] CRAN (R 4.1.2)
## ALDEx2 1.30.0 2022-11-01 [2] Bioconductor
## annotate 1.72.0 2021-10-26 [2] Bioconductor
## AnnotationDbi 1.60.2 2023-03-10 [2] Bioconductor
## ape * 5.7-1 2023-03-13 [2] CRAN (R 4.1.2)
## askpass 1.1 2019-01-13 [2] CRAN (R 4.1.0)
## backports 1.4.1 2021-12-13 [2] CRAN (R 4.1.0)
## base64enc 0.1-3 2015-07-28 [2] CRAN (R 4.1.0)
## bayesm 3.1-5 2022-12-02 [2] CRAN (R 4.1.2)
## Biobase 2.54.0 2021-10-26 [2] Bioconductor
## BiocGenerics 0.40.0 2021-10-26 [2] Bioconductor
## BiocParallel 1.28.3 2021-12-09 [2] Bioconductor
## biomformat 1.22.0 2021-10-26 [2] Bioconductor
## Biostrings 2.62.0 2021-10-26 [2] Bioconductor
## bit 4.0.5 2022-11-15 [2] CRAN (R 4.1.2)
## bit64 4.0.5 2020-08-30 [2] CRAN (R 4.1.0)
## bitops 1.0-7 2021-04-24 [2] CRAN (R 4.1.0)
## blob 1.2.4 2023-03-17 [2] CRAN (R 4.1.2)
## bookdown 0.34 2023-05-09 [2] CRAN (R 4.1.2)
## broom 1.0.5 2023-06-09 [2] CRAN (R 4.1.3)
## bslib 0.6.0 2023-11-21 [1] CRAN (R 4.1.3)
## cachem 1.0.8 2023-05-01 [2] CRAN (R 4.1.2)
## callr 3.7.3 2022-11-02 [2] CRAN (R 4.1.2)
## car 3.1-2 2023-03-30 [2] CRAN (R 4.1.2)
## carData 3.0-5 2022-01-06 [2] CRAN (R 4.1.2)
## caTools 1.18.2 2021-03-28 [2] CRAN (R 4.1.0)
## checkmate 2.2.0 2023-04-27 [2] CRAN (R 4.1.2)
## class 7.3-22 2023-05-03 [2] CRAN (R 4.1.2)
## classInt 0.4-9 2023-02-28 [2] CRAN (R 4.1.2)
## cli 3.6.1 2023-03-23 [2] CRAN (R 4.1.2)
## cluster 2.1.4 2022-08-22 [2] CRAN (R 4.1.2)
## coda 0.19-4 2020-09-30 [2] CRAN (R 4.1.0)
## codetools 0.2-19 2023-02-01 [2] CRAN (R 4.1.2)
## coin 1.4-2 2021-10-08 [2] CRAN (R 4.1.0)
## colorspace 2.1-0 2023-01-23 [2] CRAN (R 4.1.2)
## compositions 2.0-6 2023-04-13 [2] CRAN (R 4.1.2)
## conflicted * 1.2.0 2023-02-01 [2] CRAN (R 4.1.2)
## corrplot 0.92 2021-11-18 [2] CRAN (R 4.1.0)
## cowplot 1.1.1 2020-12-30 [2] CRAN (R 4.1.0)
## crayon 1.5.2 2022-09-29 [2] CRAN (R 4.1.2)
## crosstalk 1.2.0 2021-11-04 [2] CRAN (R 4.1.0)
## data.table 1.14.8 2023-02-17 [2] CRAN (R 4.1.2)
## DBI 1.1.3 2022-06-18 [2] CRAN (R 4.1.2)
## DelayedArray 0.20.0 2021-10-26 [2] Bioconductor
## DEoptimR 1.0-14 2023-06-09 [2] CRAN (R 4.1.3)
## DESeq2 1.34.0 2021-10-26 [2] Bioconductor
## devtools 2.4.5 2022-10-11 [2] CRAN (R 4.1.2)
## digest 0.6.33 2023-07-07 [1] CRAN (R 4.1.3)
## dplyr * 1.1.2 2023-04-20 [2] CRAN (R 4.1.2)
## DT 0.28 2023-05-18 [2] CRAN (R 4.1.3)
## e1071 1.7-13 2023-02-01 [2] CRAN (R 4.1.2)
## edgeR 3.36.0 2021-10-26 [2] Bioconductor
## ellipsis 0.3.2 2021-04-29 [2] CRAN (R 4.1.0)
## emmeans 1.8.7 2023-06-23 [1] CRAN (R 4.1.3)
## estimability 1.4.1 2022-08-05 [2] CRAN (R 4.1.2)
## evaluate 0.21 2023-05-05 [2] CRAN (R 4.1.2)
## FactoMineR 2.8 2023-03-27 [2] CRAN (R 4.1.2)
## fansi 1.0.4 2023-01-22 [2] CRAN (R 4.1.2)
## farver 2.1.1 2022-07-06 [2] CRAN (R 4.1.2)
## fastmap 1.1.1 2023-02-24 [2] CRAN (R 4.1.2)
## flashClust 1.01-2 2012-08-21 [2] CRAN (R 4.1.0)
## foreach 1.5.2 2022-02-02 [2] CRAN (R 4.1.2)
## foreign 0.8-84 2022-12-06 [2] CRAN (R 4.1.2)
## Formula 1.2-5 2023-02-24 [2] CRAN (R 4.1.2)
## fs 1.6.2 2023-04-25 [2] CRAN (R 4.1.2)
## genefilter 1.76.0 2021-10-26 [2] Bioconductor
## geneplotter 1.72.0 2021-10-26 [2] Bioconductor
## generics 0.1.3 2022-07-05 [2] CRAN (R 4.1.2)
## GenomeInfoDb 1.30.1 2022-01-30 [2] Bioconductor
## GenomeInfoDbData 1.2.7 2022-03-09 [2] Bioconductor
## GenomicRanges 1.46.1 2021-11-18 [2] Bioconductor
## ggiraph 0.8.7 2023-03-17 [2] CRAN (R 4.1.2)
## ggiraphExtra 0.3.0 2020-10-06 [2] CRAN (R 4.1.2)
## ggplot2 * 3.4.2 2023-04-03 [2] CRAN (R 4.1.2)
## ggpubr * 0.6.0 2023-02-10 [2] CRAN (R 4.1.2)
## ggrepel 0.9.3 2023-02-03 [2] CRAN (R 4.1.2)
## ggsci 3.0.0 2023-03-08 [2] CRAN (R 4.1.2)
## ggsignif 0.6.4 2022-10-13 [2] CRAN (R 4.1.2)
## ggVennDiagram 1.2.2 2022-09-08 [2] CRAN (R 4.1.2)
## glmnet 4.1-7 2023-03-23 [2] CRAN (R 4.1.2)
## glue 1.6.2 2022-02-24 [2] CRAN (R 4.1.2)
## gplots 3.1.3 2022-04-25 [2] CRAN (R 4.1.2)
## gridExtra 2.3 2017-09-09 [2] CRAN (R 4.1.0)
## gtable 0.3.3 2023-03-21 [2] CRAN (R 4.1.2)
## gtools 3.9.4 2022-11-27 [2] CRAN (R 4.1.2)
## highr 0.10 2022-12-22 [2] CRAN (R 4.1.2)
## Hmisc 5.1-0 2023-05-08 [2] CRAN (R 4.1.2)
## htmlTable 2.4.1 2022-07-07 [2] CRAN (R 4.1.2)
## htmltools 0.5.7 2023-11-03 [1] CRAN (R 4.1.3)
## htmlwidgets 1.6.2 2023-03-17 [2] CRAN (R 4.1.2)
## httpuv 1.6.11 2023-05-11 [2] CRAN (R 4.1.3)
## httr 1.4.6 2023-05-08 [2] CRAN (R 4.1.2)
## igraph 1.5.0 2023-06-16 [1] CRAN (R 4.1.3)
## insight 0.19.3 2023-06-29 [2] CRAN (R 4.1.3)
## IRanges 2.28.0 2021-10-26 [2] Bioconductor
## iterators 1.0.14 2022-02-05 [2] CRAN (R 4.1.2)
## jquerylib 0.1.4 2021-04-26 [2] CRAN (R 4.1.0)
## jsonlite 1.8.7 2023-06-29 [2] CRAN (R 4.1.3)
## kableExtra 1.3.4 2021-02-20 [2] CRAN (R 4.1.2)
## KEGGREST 1.34.0 2021-10-26 [2] Bioconductor
## KernSmooth 2.23-22 2023-07-10 [2] CRAN (R 4.1.3)
## knitr 1.43 2023-05-25 [2] CRAN (R 4.1.3)
## labeling 0.4.2 2020-10-20 [2] CRAN (R 4.1.0)
## later 1.3.1 2023-05-02 [2] CRAN (R 4.1.2)
## lattice * 0.21-8 2023-04-05 [2] CRAN (R 4.1.2)
## leaps 3.1 2020-01-16 [2] CRAN (R 4.1.0)
## libcoin 1.0-9 2021-09-27 [2] CRAN (R 4.1.0)
## lifecycle 1.0.3 2022-10-07 [2] CRAN (R 4.1.2)
## limma 3.50.3 2022-04-07 [2] Bioconductor
## locfit 1.5-9.8 2023-06-11 [2] CRAN (R 4.1.3)
## LOCOM 1.1 2022-08-05 [2] Github (yijuanhu/LOCOM@c181e0f)
## magrittr 2.0.3 2022-03-30 [2] CRAN (R 4.1.2)
## MASS 7.3-60 2023-05-04 [2] CRAN (R 4.1.2)
## Matrix 1.6-0 2023-07-08 [2] CRAN (R 4.1.3)
## MatrixGenerics 1.6.0 2021-10-26 [2] Bioconductor
## matrixStats 1.0.0 2023-06-02 [2] CRAN (R 4.1.3)
## mbzinb 0.2 2022-03-16 [2] local
## memoise 2.0.1 2021-11-26 [2] CRAN (R 4.1.0)
## metagenomeSeq 1.36.0 2021-10-26 [2] Bioconductor
## mgcv 1.8-42 2023-03-02 [2] CRAN (R 4.1.2)
## microbiome 1.16.0 2021-10-26 [2] Bioconductor
## mime 0.12 2021-09-28 [2] CRAN (R 4.1.0)
## miniUI 0.1.1.1 2018-05-18 [2] CRAN (R 4.1.0)
## modeltools 0.2-23 2020-03-05 [2] CRAN (R 4.1.0)
## multcomp 1.4-25 2023-06-20 [2] CRAN (R 4.1.3)
## multcompView 0.1-9 2023-04-09 [2] CRAN (R 4.1.2)
## multtest 2.50.0 2021-10-26 [2] Bioconductor
## munsell 0.5.0 2018-06-12 [2] CRAN (R 4.1.0)
## mvtnorm 1.2-2 2023-06-08 [2] CRAN (R 4.1.3)
## mycor 0.1.1 2018-04-10 [2] CRAN (R 4.1.0)
## NADA 1.6-1.1 2020-03-22 [2] CRAN (R 4.1.0)
## nlme * 3.1-162 2023-01-31 [2] CRAN (R 4.1.2)
## nnet 7.3-19 2023-05-03 [2] CRAN (R 4.1.2)
## openssl 2.0.6 2023-03-09 [2] CRAN (R 4.1.2)
## permute * 0.9-7 2022-01-27 [2] CRAN (R 4.1.2)
## pheatmap 1.0.12 2019-01-04 [2] CRAN (R 4.1.0)
## phyloseq * 1.38.0 2021-10-26 [2] Bioconductor
## picante * 1.8.2 2020-06-10 [2] CRAN (R 4.1.0)
## pillar 1.9.0 2023-03-22 [2] CRAN (R 4.1.2)
## pkgbuild 1.4.2 2023-06-26 [2] CRAN (R 4.1.3)
## pkgconfig 2.0.3 2019-09-22 [2] CRAN (R 4.1.0)
## pkgload 1.3.2.1 2023-07-08 [2] CRAN (R 4.1.3)
## plyr 1.8.8 2022-11-11 [2] CRAN (R 4.1.2)
## png 0.1-8 2022-11-29 [2] CRAN (R 4.1.2)
## ppcor 1.1 2015-12-03 [2] CRAN (R 4.1.0)
## prettyunits 1.1.1 2020-01-24 [2] CRAN (R 4.1.0)
## processx 3.8.2 2023-06-30 [2] CRAN (R 4.1.3)
## profvis 0.3.8 2023-05-02 [2] CRAN (R 4.1.2)
## promises 1.2.0.1 2021-02-11 [2] CRAN (R 4.1.0)
## protoclust 1.6.4 2022-04-01 [2] CRAN (R 4.1.2)
## proxy 0.4-27 2022-06-09 [2] CRAN (R 4.1.2)
## ps 1.7.5 2023-04-18 [2] CRAN (R 4.1.2)
## pscl 1.5.5.1 2023-05-10 [2] CRAN (R 4.1.2)
## purrr 1.0.1 2023-01-10 [2] CRAN (R 4.1.2)
## qvalue 2.26.0 2021-10-26 [2] Bioconductor
## R6 2.5.1 2021-08-19 [2] CRAN (R 4.1.0)
## RAIDA 1.0 2022-03-14 [2] local
## RColorBrewer * 1.1-3 2022-04-03 [2] CRAN (R 4.1.2)
## Rcpp 1.0.11 2023-07-06 [1] CRAN (R 4.1.3)
## RcppZiggurat 0.1.6 2020-10-20 [2] CRAN (R 4.1.0)
## RCurl 1.98-1.12 2023-03-27 [2] CRAN (R 4.1.2)
## remotes 2.4.2 2021-11-30 [2] CRAN (R 4.1.0)
## reshape2 1.4.4 2020-04-09 [2] CRAN (R 4.1.0)
## reticulate 1.30 2023-06-09 [2] CRAN (R 4.1.3)
## Rfast 2.0.8 2023-07-03 [2] CRAN (R 4.1.3)
## rhdf5 2.38.1 2022-03-10 [2] Bioconductor
## rhdf5filters 1.6.0 2021-10-26 [2] Bioconductor
## Rhdf5lib 1.16.0 2021-10-26 [2] Bioconductor
## rlang 1.1.1 2023-04-28 [1] CRAN (R 4.1.2)
## rmarkdown 2.23 2023-07-01 [2] CRAN (R 4.1.3)
## robustbase 0.99-0 2023-06-16 [2] CRAN (R 4.1.3)
## rpart 4.1.19 2022-10-21 [2] CRAN (R 4.1.2)
## RSpectra 0.16-1 2022-04-24 [2] CRAN (R 4.1.2)
## RSQLite 2.3.1 2023-04-03 [2] CRAN (R 4.1.2)
## rstatix 0.7.2 2023-02-01 [2] CRAN (R 4.1.2)
## rstudioapi 0.15.0 2023-07-07 [2] CRAN (R 4.1.3)
## Rtsne 0.16 2022-04-17 [2] CRAN (R 4.1.2)
## RVenn 1.1.0 2019-07-18 [2] CRAN (R 4.1.0)
## rvest 1.0.3 2022-08-19 [2] CRAN (R 4.1.2)
## S4Vectors 0.32.4 2022-03-29 [2] Bioconductor
## sandwich 3.0-2 2022-06-15 [2] CRAN (R 4.1.2)
## sass 0.4.6 2023-05-03 [2] CRAN (R 4.1.2)
## scales 1.2.1 2022-08-20 [2] CRAN (R 4.1.2)
## scatterplot3d 0.3-44 2023-05-05 [2] CRAN (R 4.1.2)
## sessioninfo 1.2.2 2021-12-06 [2] CRAN (R 4.1.0)
## sf 1.0-7 2022-03-07 [2] CRAN (R 4.1.2)
## shape 1.4.6 2021-05-19 [2] CRAN (R 4.1.0)
## shiny 1.7.4.1 2023-07-06 [2] CRAN (R 4.1.3)
## sjlabelled 1.2.0 2022-04-10 [2] CRAN (R 4.1.2)
## sjmisc 2.8.9 2021-12-03 [2] CRAN (R 4.1.0)
## stringi 1.7.12 2023-01-11 [2] CRAN (R 4.1.2)
## stringr 1.5.0 2022-12-02 [2] CRAN (R 4.1.2)
## SummarizedExperiment 1.24.0 2021-10-26 [2] Bioconductor
## survival 3.5-5 2023-03-12 [2] CRAN (R 4.1.2)
## svglite 2.1.1 2023-01-10 [2] CRAN (R 4.1.2)
## systemfonts 1.0.4 2022-02-11 [2] CRAN (R 4.1.2)
## tensorA 0.36.2 2020-11-19 [2] CRAN (R 4.1.0)
## TH.data 1.1-2 2023-04-17 [2] CRAN (R 4.1.2)
## tibble * 3.2.1 2023-03-20 [2] CRAN (R 4.1.2)
## tidyr 1.3.0 2023-01-24 [2] CRAN (R 4.1.2)
## tidyselect 1.2.0 2022-10-10 [2] CRAN (R 4.1.2)
## truncnorm 1.0-9 2023-03-20 [2] CRAN (R 4.1.2)
## umap 0.2.10.0 2023-02-01 [2] CRAN (R 4.1.2)
## units 0.8-2 2023-04-27 [2] CRAN (R 4.1.2)
## urlchecker 1.0.1 2021-11-30 [2] CRAN (R 4.1.0)
## usethis 2.2.2 2023-07-06 [2] CRAN (R 4.1.3)
## utf8 1.2.3 2023-01-31 [2] CRAN (R 4.1.2)
## uuid 1.1-0 2022-04-19 [2] CRAN (R 4.1.2)
## vctrs 0.6.3 2023-06-14 [1] CRAN (R 4.1.3)
## vegan * 2.6-4 2022-10-11 [2] CRAN (R 4.1.2)
## viridis * 0.6.3 2023-05-03 [2] CRAN (R 4.1.2)
## viridisLite * 0.4.2 2023-05-02 [2] CRAN (R 4.1.2)
## webshot 0.5.5 2023-06-26 [2] CRAN (R 4.1.3)
## withr 2.5.0 2022-03-03 [2] CRAN (R 4.1.2)
## Wrench 1.12.0 2021-10-26 [2] Bioconductor
## xfun 0.40 2023-08-09 [1] CRAN (R 4.1.3)
## XMAS2 * 2.2.0 2023-11-30 [1] local
## XML 3.99-0.14 2023-03-19 [2] CRAN (R 4.1.2)
## xml2 1.3.5 2023-07-06 [2] CRAN (R 4.1.3)
## xtable 1.8-4 2019-04-21 [2] CRAN (R 4.1.0)
## XVector 0.34.0 2021-10-26 [2] Bioconductor
## yaml 2.3.7 2023-01-23 [2] CRAN (R 4.1.2)
## zCompositions 1.4.0-1 2022-03-26 [2] CRAN (R 4.1.2)
## zlibbioc 1.40.0 2021-10-26 [2] Bioconductor
## zoo 1.8-12 2023-04-13 [2] CRAN (R 4.1.2)
##
## [1] /Users/zouhua/Library/R/x86_64/4.1/library
## [2] /Library/Frameworks/R.framework/Versions/4.1/Resources/library
##
## ──────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────