Chapter 2 How to install XMAS
XMAS 2.0 contains several modules, such as preprocessing, actions, methods and visualization. Furthermore, the extended parts of actions module give this package more flexible ability to add new parts. We also separate scripts into calculation and visualization parts by their names. The functions’ prefix with run is used for calculation, and the functions’ prefix with plot is used for visualization.
Outline of this Chapter:
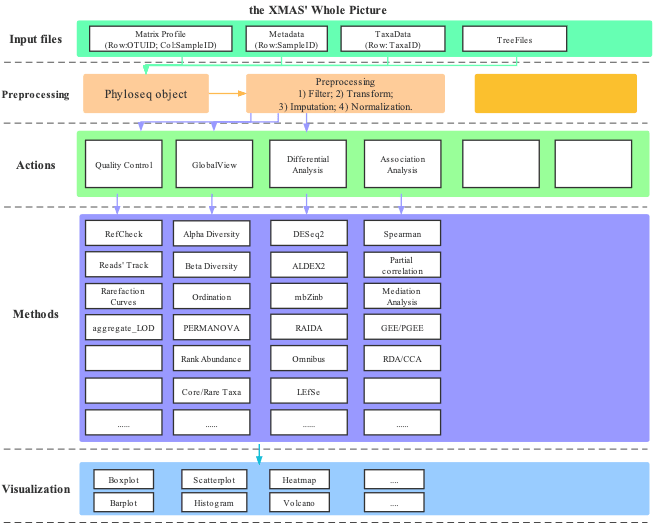
Figure 2.1: The Overview of the design of XMAS package
2.1 Description
XMAS 2.0 is comprised of R functions for in-house use, including functions for statistical, functional, and visual analysis of microbiol data, and is still underdevelopment.
Here, we provide the XMAS 2.0 releases and users can download it.
2.2 Installation of dependencies
Requiring the R-base version more than 3.6.3. Some of the dependencies are uploaded to CRAN or bioconductor website, but others, which are underdevelopment are only obtained from github website. Dependencies version (optional but not required):
- Biostrings (>= 2.22.0)
- BiocGenerics (>= 0.32.0)
- phyloseq (>= 1.30.0)
- forcats (>= 0.5.1)
- stringr (>= 1.4.0)
- ggplot2 (>= 3.3.5)
- readr (>= 1.4.0)
- ALDEx2 (>= 1.18.0)
- dplyr (>= 1.0.8)
- tibble (>= 3.1.6)
- DT (>= 0.18)
- purrr (>= 0.3.4)
- metagenomeSeq (>= 1.28.0)
- edgeR (>= 3.28.1)
- DESeq2 (>= 1.26.0)
- limma (>= 3.42.2)
- mbzinb (>= 0.2)
- pscl (>= 1.5.5)
- methods (>= 3.6.3)
- matrixStats (>= 0.58.0)
- RAIDA (>= 1.0)
- compositions (>= 2.0-4)
- vegan (>= 2.5-7)
- coin (>= 1.4-2)
- SummarizedExperiment (>= 1.16.1)
- MASS (>= 7.3-53.1)
- Biobase (>= 2.46.0)
- nlme (>= 3.1-152)
- tidyr (>= 1.1.3)
- corncob (>= 0.2.0)
- Maaslin2 (>= 1.7.3)
- ggrepel (>= 0.9.1)
- ggpubr (>= 0.4.0)
- ggalluvial (>= 0.12.3)
- Hmisc (>= 4.5-0)
- RColorBrewer (>= 1.1-2)
- umap (>= 0.2.8.0)
- ape (>= 5.6-2)
- cowplot (>= 1.1.1)
- Rtsne (>= 0.15)
- ade4 (>= 1.7-18)
- scales (>= 1.1.1)
- ggVennDiagram (>= 1.2.1)
- impute (>= 1.68.0)
- pheatmap (>= 1.0.12)
There are two methods to install the aforementioned packages.
2.2.1 Packages in CRAN & Bioconductor
install.packages("pacman")
library(pacman)
pacman::p_load(Biostrings, BiocGenerics, phyloseq, forcats, stringr, ggplot2,
readr, ALDEx2, dplyr, tibble, DT, purrr, metagenomeSeq, stats, edgeR,
DESeq2, methods, limma, pscl, matrixStats, compositions, vegan, coin,
SummarizedExperiment, MASS, Biobase, nlme, tidyr, Maaslin2, ggrepel,
ggpubr, ggalluvial, Hmisc, umap, Rtsne, ape, cowplot, scales, ade4,
ggVennDiagram, impute, pheatmap)2.2.2 Packages in github
# Step 1: Install devtools
if (!requireNamespace(c("remotes", "devtools"), quietly=TRUE)) {
install.packages(c("devtools", "remotes"))
}
library(devtools)
#library(remotes)
# Step 2: install corncob package
devtools::install_github("bryandmartin/corncob")
# remotes::install_github("bryandmartin/corncob")2.3 Installation
Get the XMAS released version from Release version
2.3.1 Installation in linux or Mac os through command line
Using git command to download the whole repository and then install this package by linux command line.
git clone git@gitlab.xbiome.com:Analytics/xmas.git
R CMD build xmas
R CMD INSTALL XMAS_*.tar.gz2.4 Vignette
For detailed tutorials on how to use XMAS, please refer to the R package vignettes. Choosing the html format to do a practice. Hope you have fun with XMAS.
2.6 Future plan
XMAS 2.0, which developed for standard data analysis workflow, is part of the X-therapia platform and we will update it with more and more data analysis modules and also add new functions according to the in-house requirements to support the company.
We welcome any comments and requirements and hope users give issues on the gitlab issues module.