Chapter 12 Association Analysis
The potential relationships between microbiota and environmental factors such as clinical parameters are vital when scientists investigate the mechanism of diseases.
Here, we identify the associations between them by using spearman correlation analysis.
Outline of this Chapter:
12.2 Importing Data
The Zeybel_Gut is from (Zeybel et al. 2022), containing gut and oral microbiota.
- sample data in Zeybel_Gut object
## Stage Metabolomics Proteomics GutMetagenomics OralMetagenomics LiverFatClass Gender AlcoholConsumption Smoker Age LF Sodium
## P101042 Before Send Send Send Send Mild Male No No 33 6.45000 142
## P101076 Before Send Send Send Send Moderate Male No No 40 9.30000 146
## P101095 Before Send Send Send Failed Sample Moderate Male No No 29 9.90000 140
## P101084 Before Send Send Send Send Severe Male No No 45 18.15000 143
## P101038 Before Send Send Send Send Moderate Female No No 43 15.93333 142
## P101071 Before Send Send Send Send Moderate Male No Yes 48 8.10000 144
## Potassium Creatinine Urea UA ALT AST GGT AP TB Albumin CK TC HDL LDL Triglycerides Glucose Insulin Weight BMI HR SBP DBP
## P101042 4.6 0.96 17 7.2 26 17 29 103 0.25 5.00 85 255 40 181 164 93 11.4 84.5 31.0 101 120 70
## P101076 4.9 1.05 22 5.6 28 16 56 81 0.34 5.00 123 193 48 133 115 111 14.7 83.0 28.4 73 120 70
## P101095 4.1 1.13 14 9.1 39 28 32 78 1.46 5.14 300 152 38 103 98 88 10.3 123.1 38.9 73 110 70
## P101084 5.1 1.15 12 8.8 57 33 55 67 0.34 4.72 336 195 47 128 124 99 36.3 108.0 33.7 74 130 80
## P101038 4.1 0.91 9 7.2 15 16 17 72 1.24 4.70 108 235 55 165 141 114 24.6 98.6 37.6 85 130 80
## P101071 4.4 1.04 19 6.0 21 13 21 70 0.34 4.60 77 228 39 146 279 104 12.5 105.8 36.6 78 110 70
## WC HC TFM TFFM TTBW LAFM LAFFM LATBW RAFM RAFFM RATBW LLFM LLFFM LLTBW RLFM RLFFM RLTBW
## P101042 100 109 13.9 32.0 44.8 1.3 3.6 44.8 1.2 3.6 44.8 3.4 10.9 44.8 3.4 11.1 44.8
## P101076 95 100 11.4 34.4 46.4 1.0 3.9 46.4 1.0 3.8 46.4 3.1 10.6 46.4 3.3 10.6 46.4
## P101095 118 128 23.8 43.2 61.5 2.7 5.0 61.5 2.3 5.1 61.5 4.8 15.5 61.5 5.5 15.2 61.5
## P101084 115 120 21.3 38.7 53.4 2.0 4.4 53.4 1.8 4.4 53.4 5.1 12.7 53.4 5.0 12.8 53.4
## P101038 100 124 23.4 28.9 52.0 3.1 2.9 52.0 2.9 2.8 52.0 8.6 8.7 52.0 8.7 8.7 52.0
## P101071 115 115 20.6 37.2 51.7 1.9 4.4 51.7 1.7 4.4 51.7 5.4 12.3 51.7 5.6 12.2 51.7## [1] "Stage" "Metabolomics" "Proteomics" "GutMetagenomics" "OralMetagenomics" "LiverFatClass"
## [7] "Gender" "AlcoholConsumption" "Smoker" "Age" "LF" "Sodium"
## [13] "Potassium" "Creatinine" "Urea" "UA" "ALT" "AST"
## [19] "GGT" "AP" "TB" "Albumin" "CK" "TC"
## [25] "HDL" "LDL" "Triglycerides" "Glucose" "Insulin" "Weight"
## [31] "BMI" "HR" "SBP" "DBP" "WC" "HC"
## [37] "TFM" "TFFM" "TTBW" "LAFM" "LAFFM" "LATBW"
## [43] "RAFM" "RAFFM" "RATBW" "LLFM" "LLFFM" "LLTBW"
## [49] "RLFM" "RLFFM" "RLTBW"- otu table profile in Zeybel_Gut object
## P101042 P101076 P101095 P101084 P101038 P101071 P101047 P101012 P101027 P101024 P101057 P101067
## g__Absiella 0.0000000 0.0000000 0.00e+00 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.00e+00 0.0000000 0.0000000 0.0000000
## g__Acidaminococcus 0.0000000 0.0012421 0.00e+00 0.0114403 0.0000000 0.0017583 0.0000000 0.0000000 0.00e+00 0.0000000 0.0000000 0.0132178
## g__Actinomyces 0.0000893 0.0003643 2.79e-05 0.0000000 0.0000000 0.0000926 0.0002638 0.0004858 0.00e+00 0.0000555 0.0006398 0.0002071
## g__Adlercreutzia 0.0000000 0.0000000 0.00e+00 0.0002282 0.0000137 0.0000000 0.0000792 0.0003161 4.06e-05 0.0001708 0.0000000 0.0000000
## g__Agathobaculum 0.0004688 0.0000000 1.84e-04 0.0006843 0.0001106 0.0000364 0.0006443 0.0000074 9.73e-05 0.0028784 0.0002393 0.0000000
## g__Aggregatibacter 0.0000000 0.0000000 0.00e+00 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.00e+00 0.0000000 0.0000000 0.0000000
## P101094 P101007 P101054 P101031 P101003 P101018 P101025 P101010 P101069 P101077 P101065 P101096
## g__Absiella 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0002135 0.00e+00 0.0000000 0.0000000 0.0000000 0.0000000
## g__Acidaminococcus 0.0000000 0.0159577 0.0012000 0.0000000 0.0000000 0.0000000 0.0009027 0.00e+00 0.0001482 0.0000000 0.0000000 0.0000000
## g__Actinomyces 0.0000000 0.0002216 0.0000024 0.0313157 0.0000000 0.0000000 0.0000000 0.00e+00 0.0000401 0.0002101 0.0001259 0.0000000
## g__Adlercreutzia 0.0001777 0.0000000 0.0000245 0.0000000 0.0000000 0.0008641 0.0000000 0.00e+00 0.0000158 0.0001580 0.0000000 0.0004234
## g__Agathobaculum 0.0020933 0.0001628 0.0007184 0.0000000 0.0002355 0.0000000 0.0061141 1.29e-05 0.0002268 0.0007352 0.0004899 0.0007549
## g__Aggregatibacter 0.0000000 0.0000000 0.0000000 0.0152900 0.0000000 0.0000000 0.0000000 0.00e+00 0.0000000 0.0000000 0.0000000 0.0000000
## P101085 P101021 P101052 P101030 P101041 P101078 P101051 P101068 P101056 P101079 P101022 P101074
## g__Absiella 0.0000000 0.0000000 0.00e+00 0.0000000 0.0000000 0.0000000 0.0000000 0.00e+00 0.00e+00 0.0000000 0.0000000 0.0000000
## g__Acidaminococcus 0.0000000 0.0000000 0.00e+00 0.0000000 0.0002144 0.0000000 0.0000000 0.00e+00 0.00e+00 0.0000000 0.0000000 0.0141640
## g__Actinomyces 0.0000000 0.0000000 1.44e-05 0.0000000 0.0000000 0.0000940 0.0000000 1.99e-05 0.00e+00 0.0000000 0.0001212 0.0000113
## g__Adlercreutzia 0.0000000 0.0000000 0.00e+00 0.0003256 0.0000000 0.0000802 0.0000000 0.00e+00 0.00e+00 0.0000000 0.0000000 0.0004728
## g__Agathobaculum 0.0020973 0.0001715 0.00e+00 0.0021537 0.0000000 0.0006245 0.0001612 2.74e-05 2.83e-05 0.0008838 0.0005000 0.0000000
## g__Aggregatibacter 0.0000000 0.0000000 0.00e+00 0.0000000 0.0000000 0.0000000 0.0000000 0.00e+00 0.00e+00 0.0000000 0.0000000 0.0000000
## P101059 P101050 P101088 P101061 P101082 P101064
## g__Absiella 0.0000000 0 0.0000000 0.0000000 0.0000000 0.0000000
## g__Acidaminococcus 0.0029344 0 0.0000000 0.0000000 0.0030215 0.0000000
## g__Actinomyces 0.0000000 0 0.0000000 0.0000299 0.0000174 0.0000093
## g__Adlercreutzia 0.0000379 0 0.0002421 0.0000383 0.0000397 0.0000200
## g__Agathobaculum 0.0025444 0 0.0022848 0.0005402 0.0013902 0.0011816
## g__Aggregatibacter 0.0000000 0 0.0000000 0.0000000 0.0000000 0.0000000Results:
- metadata has 42 continuous variables
12.3 Spearman Correlation Analysis
To identify the association between individual genus and continuous variables, we perform the correlation analysis with “spearman”, “pearson” and “kendall” method to calculate the test results. Here, the results have four statistical indexes: statistical, Rho, Pvalue and AdjustedPvalue and we also provide the plot_correlation_heatmap to display the results.
- Calculation
dat_cor <- run_cor(ps = Zeybel_Gut_genus,
columns = c("LF", "Sodium", "Potassium", "Creatinine", "Urea", "RLTBW"),
method = "spearman")
head(dat_cor)## Phenotype TaxaID Statistic Rho Pvalue AdjustedPvalue
## 1 LF g__Absiella 12102.49 0.01932697 0.9033074 0.9808251
## 2 LF g__Acidaminococcus 11509.50 0.06737739 0.6715949 0.9297863
## 3 LF g__Actinomyces 11349.08 0.08037634 0.6128579 0.9297863
## 4 LF g__Adlercreutzia 11028.15 0.10638089 0.5025215 0.9297863
## 5 LF g__Agathobaculum 13752.81 -0.11440021 0.4706651 0.9297863
## 6 LF g__Aggregatibacter 13533.57 -0.09663486 0.5426616 0.9297863- visualization
plot_correlation_heatmap(
data = dat_cor,
x_index = "Rho",
x_index_cutoff = 0,
y_index = "Pvalue",
y_index_cutoff = 0.05,
cellwidth = 35,
cellheight = 10,
fontsize_number = 15)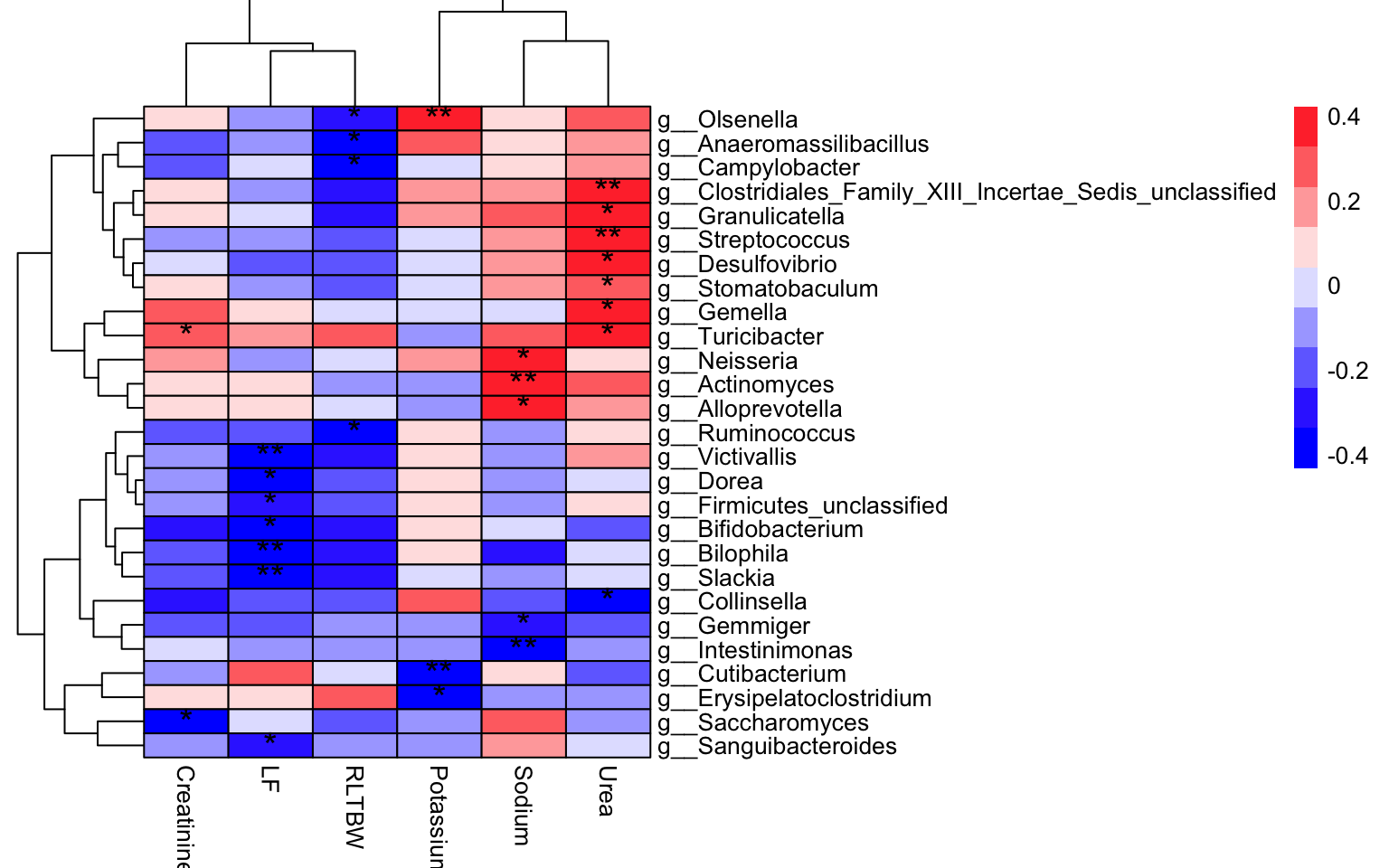
Figure 12.1: Spearman Correlation Coefficient
Results:
- the color of cell shows the size of Rho.
red: positive
blue: negative
- the asterisk of cell shows the significance:
* for [0.05, 0.01]
** for less than 0.01
12.4 Partial Correlation Analysis
To identify the association between individual genus and continuous variables, while controlling third variables, we perform the partial correlation analysis with “spearman”, “pearson” and “kendall” method to calculate the test results. Here, the results have four statistical indexes: statistical, Rho, Pvalue and AdjustedPvalue and we also provide the plot_correlation_heatmap to display the results.
When comparing the Spearman Correlation Analysis, Partial Correlation Analysis has adjusted effects from the confounding factors as third variables. For instance, we should pay attention to the age or gender etc, which could affect the test results when we do some association analysis.
- Calculation
dat_cor_partial <- run_partial_cor(ps = Zeybel_Gut_genus,
columns = c("LF", "Sodium", "Potassium", "Creatinine", "Urea", "RLTBW"),
AdjVars = c("Age", "Gender", "Smoker"),
method_t = "pcor",
method = "spearman",
p_adjust = "BH")
head(dat_cor_partial)## Phenotype TaxaID Statistic Rho Pvalue AdjustedPvalue
## 1 LF g__Absiella 0.07849237 0.01290299 0.9378594 0.9942613
## 2 LF g__Acidaminococcus 0.41767935 0.06850475 0.6785956 0.9738542
## 3 LF g__Actinomyces 0.53412751 0.08747343 0.5964482 0.9738542
## 4 LF g__Adlercreutzia 0.54397242 0.08907305 0.5897236 0.9738542
## 5 LF g__Agathobaculum -0.43073071 -0.07063482 0.6691631 0.9738542
## 6 LF g__Aggregatibacter -0.34019586 -0.05584059 0.7356329 0.9738542- visualization
plot_correlation_heatmap(
data = dat_cor_partial,
x_index = "Rho",
x_index_cutoff = 0,
y_index = "Pvalue",
y_index_cutoff = 0.05,
cellwidth = 35,
cellheight = 10,
fontsize_number = 15)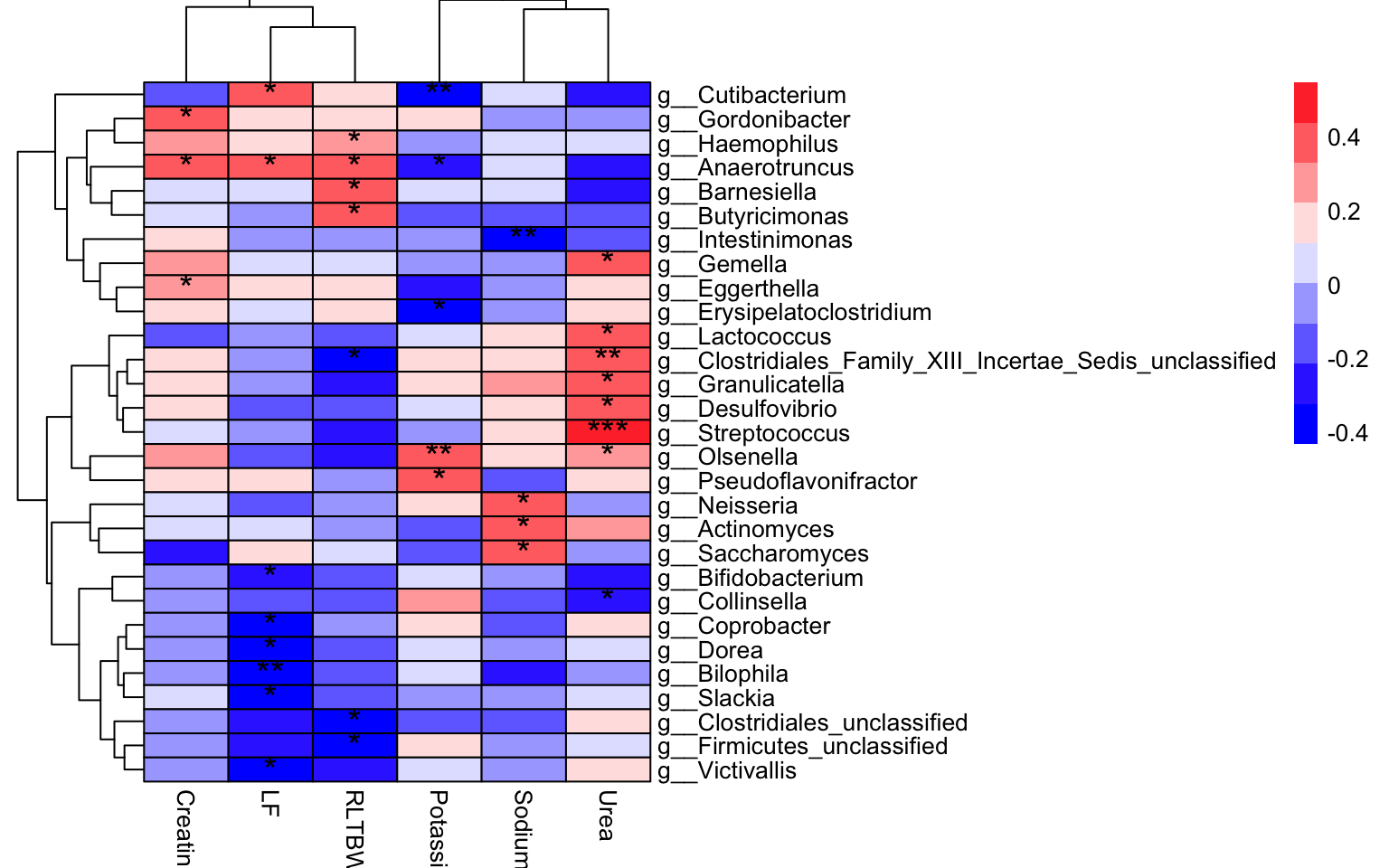
Figure 12.2: Partial Correlation Coefficient
Results:
- the color of cell shows the size of Rho.
red: positive
blue: negative
- the asterisk of cell shows the significance:
* for [0.05, 0.01]
** for less than 0.01
12.5 Systematic Information
## ─ Session info ───────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────
## setting value
## version R version 4.1.3 (2022-03-10)
## os macOS Monterey 12.2.1
## system x86_64, darwin17.0
## ui RStudio
## language (EN)
## collate en_US.UTF-8
## ctype en_US.UTF-8
## tz Asia/Shanghai
## date 2023-11-30
## rstudio 2023.09.0+463 Desert Sunflower (desktop)
## pandoc 3.1.1 @ /Applications/RStudio.app/Contents/Resources/app/quarto/bin/tools/ (via rmarkdown)
##
## ─ Packages ───────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────
## package * version date (UTC) lib source
## abind 1.4-5 2016-07-21 [2] CRAN (R 4.1.0)
## ade4 1.7-22 2023-02-06 [2] CRAN (R 4.1.2)
## ALDEx2 1.30.0 2022-11-01 [2] Bioconductor
## annotate 1.72.0 2021-10-26 [2] Bioconductor
## AnnotationDbi 1.60.2 2023-03-10 [2] Bioconductor
## ape * 5.7-1 2023-03-13 [2] CRAN (R 4.1.2)
## askpass 1.1 2019-01-13 [2] CRAN (R 4.1.0)
## backports 1.4.1 2021-12-13 [2] CRAN (R 4.1.0)
## base64enc 0.1-3 2015-07-28 [2] CRAN (R 4.1.0)
## bayesm 3.1-5 2022-12-02 [2] CRAN (R 4.1.2)
## Biobase 2.54.0 2021-10-26 [2] Bioconductor
## BiocGenerics 0.40.0 2021-10-26 [2] Bioconductor
## BiocParallel 1.28.3 2021-12-09 [2] Bioconductor
## biomformat 1.22.0 2021-10-26 [2] Bioconductor
## Biostrings 2.62.0 2021-10-26 [2] Bioconductor
## bit 4.0.5 2022-11-15 [2] CRAN (R 4.1.2)
## bit64 4.0.5 2020-08-30 [2] CRAN (R 4.1.0)
## bitops 1.0-7 2021-04-24 [2] CRAN (R 4.1.0)
## blob 1.2.4 2023-03-17 [2] CRAN (R 4.1.2)
## bookdown 0.34 2023-05-09 [2] CRAN (R 4.1.2)
## broom 1.0.5 2023-06-09 [2] CRAN (R 4.1.3)
## bslib 0.6.0 2023-11-21 [1] CRAN (R 4.1.3)
## cachem 1.0.8 2023-05-01 [2] CRAN (R 4.1.2)
## callr 3.7.3 2022-11-02 [2] CRAN (R 4.1.2)
## car 3.1-2 2023-03-30 [2] CRAN (R 4.1.2)
## carData 3.0-5 2022-01-06 [2] CRAN (R 4.1.2)
## caTools 1.18.2 2021-03-28 [2] CRAN (R 4.1.0)
## checkmate 2.2.0 2023-04-27 [2] CRAN (R 4.1.2)
## class 7.3-22 2023-05-03 [2] CRAN (R 4.1.2)
## classInt 0.4-9 2023-02-28 [2] CRAN (R 4.1.2)
## cli 3.6.1 2023-03-23 [2] CRAN (R 4.1.2)
## cluster 2.1.4 2022-08-22 [2] CRAN (R 4.1.2)
## coda 0.19-4 2020-09-30 [2] CRAN (R 4.1.0)
## codetools 0.2-19 2023-02-01 [2] CRAN (R 4.1.2)
## coin 1.4-2 2021-10-08 [2] CRAN (R 4.1.0)
## colorspace 2.1-0 2023-01-23 [2] CRAN (R 4.1.2)
## compositions 2.0-6 2023-04-13 [2] CRAN (R 4.1.2)
## conflicted * 1.2.0 2023-02-01 [2] CRAN (R 4.1.2)
## corrplot 0.92 2021-11-18 [2] CRAN (R 4.1.0)
## cowplot 1.1.1 2020-12-30 [2] CRAN (R 4.1.0)
## crayon 1.5.2 2022-09-29 [2] CRAN (R 4.1.2)
## crosstalk 1.2.0 2021-11-04 [2] CRAN (R 4.1.0)
## data.table 1.14.8 2023-02-17 [2] CRAN (R 4.1.2)
## DBI 1.1.3 2022-06-18 [2] CRAN (R 4.1.2)
## DelayedArray 0.20.0 2021-10-26 [2] Bioconductor
## DEoptimR 1.0-14 2023-06-09 [2] CRAN (R 4.1.3)
## DESeq2 1.34.0 2021-10-26 [2] Bioconductor
## devtools 2.4.5 2022-10-11 [2] CRAN (R 4.1.2)
## digest 0.6.33 2023-07-07 [1] CRAN (R 4.1.3)
## dplyr * 1.1.2 2023-04-20 [2] CRAN (R 4.1.2)
## DT 0.28 2023-05-18 [2] CRAN (R 4.1.3)
## e1071 1.7-13 2023-02-01 [2] CRAN (R 4.1.2)
## edgeR 3.36.0 2021-10-26 [2] Bioconductor
## ellipsis 0.3.2 2021-04-29 [2] CRAN (R 4.1.0)
## emmeans 1.8.7 2023-06-23 [1] CRAN (R 4.1.3)
## estimability 1.4.1 2022-08-05 [2] CRAN (R 4.1.2)
## evaluate 0.21 2023-05-05 [2] CRAN (R 4.1.2)
## FactoMineR 2.8 2023-03-27 [2] CRAN (R 4.1.2)
## fansi 1.0.4 2023-01-22 [2] CRAN (R 4.1.2)
## farver 2.1.1 2022-07-06 [2] CRAN (R 4.1.2)
## fastmap 1.1.1 2023-02-24 [2] CRAN (R 4.1.2)
## flashClust 1.01-2 2012-08-21 [2] CRAN (R 4.1.0)
## foreach 1.5.2 2022-02-02 [2] CRAN (R 4.1.2)
## foreign 0.8-84 2022-12-06 [2] CRAN (R 4.1.2)
## Formula 1.2-5 2023-02-24 [2] CRAN (R 4.1.2)
## fs 1.6.2 2023-04-25 [2] CRAN (R 4.1.2)
## genefilter 1.76.0 2021-10-26 [2] Bioconductor
## geneplotter 1.72.0 2021-10-26 [2] Bioconductor
## generics 0.1.3 2022-07-05 [2] CRAN (R 4.1.2)
## GenomeInfoDb 1.30.1 2022-01-30 [2] Bioconductor
## GenomeInfoDbData 1.2.7 2022-03-09 [2] Bioconductor
## GenomicRanges 1.46.1 2021-11-18 [2] Bioconductor
## ggiraph 0.8.7 2023-03-17 [2] CRAN (R 4.1.2)
## ggiraphExtra 0.3.0 2020-10-06 [2] CRAN (R 4.1.2)
## ggplot2 * 3.4.2 2023-04-03 [2] CRAN (R 4.1.2)
## ggpubr * 0.6.0 2023-02-10 [2] CRAN (R 4.1.2)
## ggrepel 0.9.3 2023-02-03 [2] CRAN (R 4.1.2)
## ggsci 3.0.0 2023-03-08 [2] CRAN (R 4.1.2)
## ggsignif 0.6.4 2022-10-13 [2] CRAN (R 4.1.2)
## ggVennDiagram 1.2.2 2022-09-08 [2] CRAN (R 4.1.2)
## glmnet 4.1-7 2023-03-23 [2] CRAN (R 4.1.2)
## glue 1.6.2 2022-02-24 [2] CRAN (R 4.1.2)
## gplots 3.1.3 2022-04-25 [2] CRAN (R 4.1.2)
## gridExtra 2.3 2017-09-09 [2] CRAN (R 4.1.0)
## gtable 0.3.3 2023-03-21 [2] CRAN (R 4.1.2)
## gtools 3.9.4 2022-11-27 [2] CRAN (R 4.1.2)
## highr 0.10 2022-12-22 [2] CRAN (R 4.1.2)
## Hmisc 5.1-0 2023-05-08 [2] CRAN (R 4.1.2)
## htmlTable 2.4.1 2022-07-07 [2] CRAN (R 4.1.2)
## htmltools 0.5.7 2023-11-03 [1] CRAN (R 4.1.3)
## htmlwidgets 1.6.2 2023-03-17 [2] CRAN (R 4.1.2)
## httpuv 1.6.11 2023-05-11 [2] CRAN (R 4.1.3)
## httr 1.4.6 2023-05-08 [2] CRAN (R 4.1.2)
## igraph 1.5.0 2023-06-16 [1] CRAN (R 4.1.3)
## insight 0.19.3 2023-06-29 [2] CRAN (R 4.1.3)
## IRanges 2.28.0 2021-10-26 [2] Bioconductor
## iterators 1.0.14 2022-02-05 [2] CRAN (R 4.1.2)
## jquerylib 0.1.4 2021-04-26 [2] CRAN (R 4.1.0)
## jsonlite 1.8.7 2023-06-29 [2] CRAN (R 4.1.3)
## kableExtra 1.3.4 2021-02-20 [2] CRAN (R 4.1.2)
## KEGGREST 1.34.0 2021-10-26 [2] Bioconductor
## KernSmooth 2.23-22 2023-07-10 [2] CRAN (R 4.1.3)
## knitr 1.43 2023-05-25 [2] CRAN (R 4.1.3)
## labeling 0.4.2 2020-10-20 [2] CRAN (R 4.1.0)
## later 1.3.1 2023-05-02 [2] CRAN (R 4.1.2)
## lattice * 0.21-8 2023-04-05 [2] CRAN (R 4.1.2)
## leaps 3.1 2020-01-16 [2] CRAN (R 4.1.0)
## libcoin 1.0-9 2021-09-27 [2] CRAN (R 4.1.0)
## lifecycle 1.0.3 2022-10-07 [2] CRAN (R 4.1.2)
## limma 3.50.3 2022-04-07 [2] Bioconductor
## locfit 1.5-9.8 2023-06-11 [2] CRAN (R 4.1.3)
## LOCOM 1.1 2022-08-05 [2] Github (yijuanhu/LOCOM@c181e0f)
## magrittr 2.0.3 2022-03-30 [2] CRAN (R 4.1.2)
## MASS 7.3-60 2023-05-04 [2] CRAN (R 4.1.2)
## Matrix 1.6-0 2023-07-08 [2] CRAN (R 4.1.3)
## MatrixGenerics 1.6.0 2021-10-26 [2] Bioconductor
## matrixStats 1.0.0 2023-06-02 [2] CRAN (R 4.1.3)
## mbzinb 0.2 2022-03-16 [2] local
## memoise 2.0.1 2021-11-26 [2] CRAN (R 4.1.0)
## metagenomeSeq 1.36.0 2021-10-26 [2] Bioconductor
## mgcv 1.8-42 2023-03-02 [2] CRAN (R 4.1.2)
## microbiome 1.16.0 2021-10-26 [2] Bioconductor
## mime 0.12 2021-09-28 [2] CRAN (R 4.1.0)
## miniUI 0.1.1.1 2018-05-18 [2] CRAN (R 4.1.0)
## modeltools 0.2-23 2020-03-05 [2] CRAN (R 4.1.0)
## multcomp 1.4-25 2023-06-20 [2] CRAN (R 4.1.3)
## multcompView 0.1-9 2023-04-09 [2] CRAN (R 4.1.2)
## multtest 2.50.0 2021-10-26 [2] Bioconductor
## munsell 0.5.0 2018-06-12 [2] CRAN (R 4.1.0)
## mvtnorm 1.2-2 2023-06-08 [2] CRAN (R 4.1.3)
## mycor 0.1.1 2018-04-10 [2] CRAN (R 4.1.0)
## NADA 1.6-1.1 2020-03-22 [2] CRAN (R 4.1.0)
## nlme * 3.1-162 2023-01-31 [2] CRAN (R 4.1.2)
## nnet 7.3-19 2023-05-03 [2] CRAN (R 4.1.2)
## openssl 2.0.6 2023-03-09 [2] CRAN (R 4.1.2)
## permute * 0.9-7 2022-01-27 [2] CRAN (R 4.1.2)
## pheatmap * 1.0.12 2019-01-04 [2] CRAN (R 4.1.0)
## phyloseq * 1.38.0 2021-10-26 [2] Bioconductor
## picante * 1.8.2 2020-06-10 [2] CRAN (R 4.1.0)
## pillar 1.9.0 2023-03-22 [2] CRAN (R 4.1.2)
## pkgbuild 1.4.2 2023-06-26 [2] CRAN (R 4.1.3)
## pkgconfig 2.0.3 2019-09-22 [2] CRAN (R 4.1.0)
## pkgload 1.3.2.1 2023-07-08 [2] CRAN (R 4.1.3)
## plyr 1.8.8 2022-11-11 [2] CRAN (R 4.1.2)
## png 0.1-8 2022-11-29 [2] CRAN (R 4.1.2)
## ppcor 1.1 2015-12-03 [2] CRAN (R 4.1.0)
## prettyunits 1.1.1 2020-01-24 [2] CRAN (R 4.1.0)
## processx 3.8.2 2023-06-30 [2] CRAN (R 4.1.3)
## profvis 0.3.8 2023-05-02 [2] CRAN (R 4.1.2)
## promises 1.2.0.1 2021-02-11 [2] CRAN (R 4.1.0)
## protoclust 1.6.4 2022-04-01 [2] CRAN (R 4.1.2)
## proxy 0.4-27 2022-06-09 [2] CRAN (R 4.1.2)
## ps 1.7.5 2023-04-18 [2] CRAN (R 4.1.2)
## pscl 1.5.5.1 2023-05-10 [2] CRAN (R 4.1.2)
## purrr 1.0.1 2023-01-10 [2] CRAN (R 4.1.2)
## qvalue 2.26.0 2021-10-26 [2] Bioconductor
## R6 2.5.1 2021-08-19 [2] CRAN (R 4.1.0)
## RAIDA 1.0 2022-03-14 [2] local
## RColorBrewer * 1.1-3 2022-04-03 [2] CRAN (R 4.1.2)
## Rcpp 1.0.11 2023-07-06 [1] CRAN (R 4.1.3)
## RcppZiggurat 0.1.6 2020-10-20 [2] CRAN (R 4.1.0)
## RCurl 1.98-1.12 2023-03-27 [2] CRAN (R 4.1.2)
## remotes 2.4.2 2021-11-30 [2] CRAN (R 4.1.0)
## reshape2 1.4.4 2020-04-09 [2] CRAN (R 4.1.0)
## reticulate 1.30 2023-06-09 [2] CRAN (R 4.1.3)
## Rfast 2.0.8 2023-07-03 [2] CRAN (R 4.1.3)
## rhdf5 2.38.1 2022-03-10 [2] Bioconductor
## rhdf5filters 1.6.0 2021-10-26 [2] Bioconductor
## Rhdf5lib 1.16.0 2021-10-26 [2] Bioconductor
## rlang 1.1.1 2023-04-28 [1] CRAN (R 4.1.2)
## rmarkdown 2.23 2023-07-01 [2] CRAN (R 4.1.3)
## robustbase 0.99-0 2023-06-16 [2] CRAN (R 4.1.3)
## rpart 4.1.19 2022-10-21 [2] CRAN (R 4.1.2)
## RSpectra 0.16-1 2022-04-24 [2] CRAN (R 4.1.2)
## RSQLite 2.3.1 2023-04-03 [2] CRAN (R 4.1.2)
## rstatix 0.7.2 2023-02-01 [2] CRAN (R 4.1.2)
## rstudioapi 0.15.0 2023-07-07 [2] CRAN (R 4.1.3)
## Rtsne 0.16 2022-04-17 [2] CRAN (R 4.1.2)
## RVenn 1.1.0 2019-07-18 [2] CRAN (R 4.1.0)
## rvest 1.0.3 2022-08-19 [2] CRAN (R 4.1.2)
## S4Vectors 0.32.4 2022-03-29 [2] Bioconductor
## sandwich 3.0-2 2022-06-15 [2] CRAN (R 4.1.2)
## sass 0.4.6 2023-05-03 [2] CRAN (R 4.1.2)
## scales 1.2.1 2022-08-20 [2] CRAN (R 4.1.2)
## scatterplot3d 0.3-44 2023-05-05 [2] CRAN (R 4.1.2)
## sessioninfo 1.2.2 2021-12-06 [2] CRAN (R 4.1.0)
## sf 1.0-7 2022-03-07 [2] CRAN (R 4.1.2)
## shape 1.4.6 2021-05-19 [2] CRAN (R 4.1.0)
## shiny 1.7.4.1 2023-07-06 [2] CRAN (R 4.1.3)
## sjlabelled 1.2.0 2022-04-10 [2] CRAN (R 4.1.2)
## sjmisc 2.8.9 2021-12-03 [2] CRAN (R 4.1.0)
## stringi 1.7.12 2023-01-11 [2] CRAN (R 4.1.2)
## stringr 1.5.0 2022-12-02 [2] CRAN (R 4.1.2)
## SummarizedExperiment 1.24.0 2021-10-26 [2] Bioconductor
## survival 3.5-5 2023-03-12 [2] CRAN (R 4.1.2)
## svglite 2.1.1 2023-01-10 [2] CRAN (R 4.1.2)
## systemfonts 1.0.4 2022-02-11 [2] CRAN (R 4.1.2)
## tensorA 0.36.2 2020-11-19 [2] CRAN (R 4.1.0)
## TH.data 1.1-2 2023-04-17 [2] CRAN (R 4.1.2)
## tibble * 3.2.1 2023-03-20 [2] CRAN (R 4.1.2)
## tidyr 1.3.0 2023-01-24 [2] CRAN (R 4.1.2)
## tidyselect 1.2.0 2022-10-10 [2] CRAN (R 4.1.2)
## truncnorm 1.0-9 2023-03-20 [2] CRAN (R 4.1.2)
## umap 0.2.10.0 2023-02-01 [2] CRAN (R 4.1.2)
## units 0.8-2 2023-04-27 [2] CRAN (R 4.1.2)
## urlchecker 1.0.1 2021-11-30 [2] CRAN (R 4.1.0)
## usethis 2.2.2 2023-07-06 [2] CRAN (R 4.1.3)
## utf8 1.2.3 2023-01-31 [2] CRAN (R 4.1.2)
## uuid 1.1-0 2022-04-19 [2] CRAN (R 4.1.2)
## vctrs 0.6.3 2023-06-14 [1] CRAN (R 4.1.3)
## vegan * 2.6-4 2022-10-11 [2] CRAN (R 4.1.2)
## viridis * 0.6.3 2023-05-03 [2] CRAN (R 4.1.2)
## viridisLite * 0.4.2 2023-05-02 [2] CRAN (R 4.1.2)
## webshot 0.5.5 2023-06-26 [2] CRAN (R 4.1.3)
## withr 2.5.0 2022-03-03 [2] CRAN (R 4.1.2)
## Wrench 1.12.0 2021-10-26 [2] Bioconductor
## xfun 0.40 2023-08-09 [1] CRAN (R 4.1.3)
## XMAS2 * 2.2.0 2023-11-30 [1] local
## XML 3.99-0.14 2023-03-19 [2] CRAN (R 4.1.2)
## xml2 1.3.5 2023-07-06 [2] CRAN (R 4.1.3)
## xtable 1.8-4 2019-04-21 [2] CRAN (R 4.1.0)
## XVector 0.34.0 2021-10-26 [2] Bioconductor
## yaml 2.3.7 2023-01-23 [2] CRAN (R 4.1.2)
## zCompositions 1.4.0-1 2022-03-26 [2] CRAN (R 4.1.2)
## zlibbioc 1.40.0 2021-10-26 [2] Bioconductor
## zoo 1.8-12 2023-04-13 [2] CRAN (R 4.1.2)
##
## [1] /Users/zouhua/Library/R/x86_64/4.1/library
## [2] /Library/Frameworks/R.framework/Versions/4.1/Resources/library
##
## ──────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────