Chapter 5 MSEA
This is a markdown tutorial for MSEA. To run MSEA, you need to Install MSEA to your default python before running this template. Installation instruction: https://msea.readthedocs.io/en/latest/quickstart.html#installation.
You can run MSEA when you have genus that you’re interested in (e.g., DA genus), write all genus into one txt file, one genus per line and feed the file to MSEA as input.
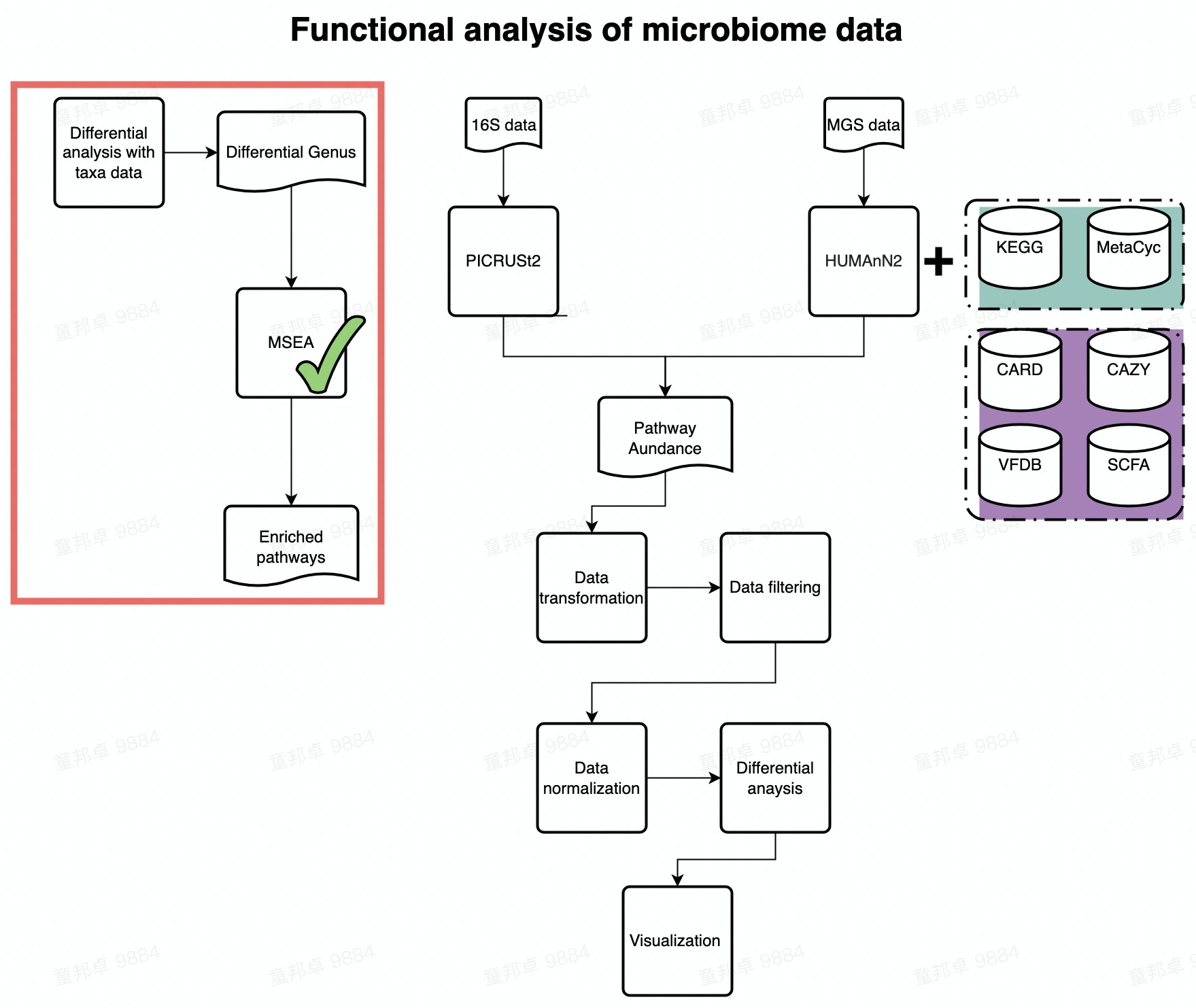
Flowchart
5.1 Execute MSEA with python script on server
You can run MSEA analysis with MSEA_Run.py on server, one required parameters and two optional parameters need to be provided for the script:
- –input (Required), input genus list file for MSEA, one genus per line.
- –output (Optional), output csv file of MSEA result. Default testout.csv
- –PerturbationTimes (Optional), number of perturbation. Fisher’s excat test has bias on group with large number of samples, MSEA uses random sampling to remove the bias. Larger number of perturbation would cause longer runtime. Default 50.
For more detail, see citation
/home/tongbangzhuo/Software/miniconda3/bin/python ./MSEA/MSEA_Run.py --input ./MSEA/test_input --output ./MSEA/testout.csv --PerturbationTimes 105.2 Environment setup
5.3 Read in MSEA result
MSEA_res = read.csv('/share/projects/SOP/Functional_Analysis/github/Functional_analysis/MSEA/testout.csv', sep = '\t')
head(MSEA_res, n=3)## term oddsratio pvalue qvalue zscore combined_score shared n_shared
## 1 SP7 27.58621 9.571517e-05 0.0003983486 -8.376409 77.51640 ['Pseudomonas', 'Salmonella', 'Azomonas', 'Sodalis'] 4
## 2 SOCS3 29.41176 1.234368e-05 0.0001439254 -5.916120 66.86615 ['Salmonella', 'Azomonas', 'Sodalis', 'Pseudomonas', 'Borrelia'] 5
## 3 IFNAR1 53.33333 1.010645e-05 0.0001312818 -5.152460 59.26533 ['Pseudomonas', 'Salmonella', 'Borrelia', 'Sodalis'] 4As the table shown above, MSEA result has 8 columns:
- term, human gene names.
- oddsratio. Odds ratio (Effect size) of the association between human gene and microbial Genus.
- pvalue. p value of Fisher’s exact test.
- qvalue. q value of Fisher’s exact test. FDR Benjamini-Hochberg correction applied.
- zscore. z-score measuring the deviation in expected ranks.
- combined_score. \(c = log_{10}(p)*z\).
- shared. Genus associated with the human gene.
- n_shared. Number of genus associated with the human gene.
5.4 Filter MSEA result
## Filter MSEA result with qvalue
MSEA_res %<>% filter(qvalue < 0.05)
## Draw bipartite with top 10 combined_score human genes
## Define data transforming function
Transform_data <- function(df){
All_Genus_in_res <- df$shared %>% unlist() %>% str_remove_all('\\[') %>% str_remove_all('\\]') %>% str_remove_all("'") %>% str_split(', ') %>% unlist() %>% unique()
lst <- list()
for (i in All_Genus_in_res){
count_vec <- c()
for (j in (1:nrow(df))){
target_string = df[j,'shared'] %>% str_remove_all('\\[') %>% str_remove_all('\\]') %>% str_remove_all("'") %>% str_split(', ') %>% unlist()
count = sum(i == target_string)
count_vec <- c(count_vec, count)
}
lst[[i]] <- count_vec
}
bipartite_tbl <- cbind(df, as.data.frame(lst)) %>% dplyr::select(term, all_of(All_Genus_in_res)) %>% column_to_rownames('term')
return(bipartite_tbl)
}
## Select top 10 human genes
Top_MSEA_res <- MSEA_res[1:10,]5.5 Visualization
Draw bipartit graph to show the relation between human genes and microbial Genus with ggnet.
## Generate data for bipartit graph
bipartite_tbl <- Transform_data(Top_MSEA_res)
## Define network layout
mymat <- bipartite_tbl
coordP <- cbind(rep(2, dim(mymat)[1]), seq(1, dim(mymat)[1]) +
2)
coordA <- cbind(rep(4, dim(mymat)[2]), seq(1, dim(mymat)[2]) +
2)
mylayout <- as.matrix(rbind(coordP, coordA))
## Construct network content
test.net <- bip_init_network(mymat, mode1 = 'HumanGenes',mode2 = 'MicrobialGenus')
# Define groups of network nodes
test.net %v% "Group" = get.vertex.attribute(test.net, attrname="mode")
# Draw network
p <- GGally::ggnet2(test.net, mode = mylayout,
label = T, size = "degree",
color = 'Group', shape = 'Group',
label.size = 5, layout.exp = 1.5, alpha = 0.75) +
scale_colour_manual(values = wes_palette("FantasticFox1")) +
guides(color=guide_legend("Group"))
p
5.6 Run enrichR analysis
After acquiring genus-associated human genes, you can run enrich your genes on different databases by enrichR.
The next chunk shows you how to run enrichR on R studio, you can either run enrichR on their interactive website.
library(enrichR)
## List available types of databases
listEnrichrSites()
## Choose database of Human genes
setEnrichrSite("Enrichr")
## List available database
websiteLive <- TRUE
dbs <- listEnrichrDbs()
if (is.null(dbs)) websiteLive <- FALSE
if (websiteLive) head(dbs)## geneCoverage genesPerTerm libraryName link numTerms appyter
## 1 13362 275 Genome_Browser_PWMs http://hgdownload.cse.ucsc.edu/goldenPath/hg18/database/ 615 ea115789fcbf12797fd692cec6df0ab4dbc79c6a
## 2 27884 1284 TRANSFAC_and_JASPAR_PWMs http://jaspar.genereg.net/html/DOWNLOAD/ 326 7d42eb43a64a4e3b20d721fc7148f685b53b6b30
## 3 6002 77 Transcription_Factor_PPIs 290 849f222220618e2599d925b6b51868cf1dab3763
## 4 47172 1370 ChEA_2013 http://amp.pharm.mssm.edu/lib/cheadownload.jsp 353 7ebe772afb55b63b41b79dd8d06ea0fdd9fa2630
## 5 47107 509 Drug_Perturbations_from_GEO_2014 http://www.ncbi.nlm.nih.gov/geo/ 701 ad270a6876534b7cb063e004289dcd4d3164f342
## 6 21493 3713 ENCODE_TF_ChIP-seq_2014 http://genome.ucsc.edu/ENCODE/downloads.html 498 497787ebc418d308045efb63b8586f10c526af51
## categoryId
## 1 1
## 2 1
## 3 1
## 4 7
## 5 7
## 6 7## Choose the databases you want to enrich your genes with and Run enrichR of genus-associated human genes on chosen databases
dbs <- c("GO_Molecular_Function_2021","KEGG_2019_Human")
if (websiteLive) {
enriched <- enrichr(MSEA_res$term %>% as.vector(), dbs)
}## Uploading data to Enrichr... Done.
## Querying GO_Molecular_Function_2021... Done.
## Querying KEGG_2019_Human... Done.
## Parsing results... Done.## Show first few rows of enrichment result
Kegg_res <- enriched[['KEGG_2019_Human']]
head(Kegg_res)## Term Overlap P.value Adjusted.P.value Old.P.value Old.Adjusted.P.value Odds.Ratio Combined.Score
## 1 Pathways in cancer 118/530 3.368482e-53 6.029198e-51 0 0 7.523619 909.0228
## 2 Cytokine-cytokine receptor interaction 90/294 4.368984e-53 6.029198e-51 0 0 11.275567 1359.4103
## 3 IL-17 signaling pathway 52/93 2.802254e-47 2.578073e-45 0 0 31.100813 3333.7296
## 4 Hepatitis B 63/163 2.226240e-44 1.536105e-42 0 0 15.621378 1570.1583
## 5 Kaposi sarcoma-associated herpesvirus infection 66/186 1.153286e-43 6.366139e-42 0 0 13.676762 1352.2016
## 6 Hepatitis C 61/155 1.497013e-43 6.886260e-42 0 0 16.054226 1583.0701
## Genes
## 1 SPI1;EPO;ARAF;KEAP1;FGF2;CRKL;FGF7;CCND1;CDH1;MYC;AKT2;AKT1;SKP1;MAP2K1;HGF;WNT5A;MITF;PGF;RUNX1;TP53;IFNAR1;NOTCH1;PDGFB;HIF1A;BCL2L11;TERT;ABL1;HMOX1;PMAIP1;FADD;SMAD2;TGFB2;SMAD4;GSTM1;TGFB1;SMAD3;TGFB3;WNT3A;NFKB1;PTK2;IL2;IL4;NFKBIA;BMP4;BMP2;IL6;CXCL12;CDK6;CDK4;CDK2;GNAS;GRB2;FGFR2;BCL2L1;NFE2L2;FGFR1;RET;ALK;CDKN1A;CXCL8;FLT3;PTEN;SLC2A1;FASLG;BRCA2;IKBKB;CASP9;CASP7;CASP8;CASP3;RAC2;JAK2;HRAS;APAF1;CHUK;IFNGR1;IL15;MMP1;MMP2;IL13;FOS;TRAF1;MMP9;RHOA;IFNG;TRAF6;KIT;CRK;CEBPA;RALA;HDAC1;GSTP1;XIAP;CXCR4;PTGS2;CBL;DLL1;EGFR;RELA;CDC42;MAPK8;ERBB2;STAT4;STAT6;BAK1;IL12RB1;NQO1;NOS2;STAT1;EGF;STAT2;STAT3;MTOR;RAD51;CTNNB1;FAS;BAX;KRAS
## 2 IL21;CXCL6;IL1RN;CD40;CSF3;CXCL9;CSF2;EDA;CXCL8;EPO;FASLG;CXCL1;PRL;CXCL13;CXCL3;TNF;CXCL2;CX3CL1;CXCL5;TNFSF13B;CCR9;TNFSF11;CCR7;CCR5;AMH;CCR3;CCR2;IL10;IL11;IL15;IFNGR1;IL13;IL18;NGF;IL17RA;TNFRSF1A;IL1A;IFNG;IL1B;LTB;IFNAR1;CX3CR1;CCL13;CCL11;CXCR4;TNFRSF11B;CXCR6;CXCR1;CCL7;CXCR3;CXCR2;CCL2;CCL1;CCL19;IL12RB1;CCL17;CCR1;CCL25;IL33;TGFB2;CCL22;TGFB1;TSLP;CCL21;TNFSF15;CCL20;TGFB3;LIF;PPBP;BMP7;IL2;IL4;BMP4;GH2;CXCL10;CXCL11;BMP2;IL6;CD4;CXCL12;LEP;CD27;FAS;IL17F;LTBR;INHA;IL17C;CCL27;IL17A;PF4
## 3 CXCL6;CSF3;CXCL8;CSF2;TRADD;TNFAIP3;CXCL1;CXCL3;CXCL2;TNF;CXCL5;IKBKB;CASP8;TBK1;CASP3;CHUK;MMP1;MMP3;IL13;FOS;MMP9;IL17RA;MMP13;IFNG;IL1B;TRAF6;DEFB4A;S100A9;S100A7;CEBPB;CCL11;PTGS2;RELA;MAPK8;CCL7;CCL2;FADD;CCL17;JUND;CCL20;MAPK14;MUC5B;NFKB1;IL4;NFKBIA;CXCL10;IL6;LCN2;FOSB;IL17F;IL17C;IL17A
## 4 CDKN1A;CXCL8;ARAF;FASLG;TNF;CASP9;IKBKB;TBK1;CASP8;MYC;AKT2;CASP3;AKT1;JAK2;HRAS;MAP2K1;APAF1;CHUK;DDX58;IRAK4;TYK2;FOS;MMP9;TIRAP;CCNA2;CREB1;IRF3;TRAF6;IRF7;TP53;TLR4;TLR3;IFNAR1;ATF4;TLR2;SRC;RELA;MAPK8;IRAK1;STAT4;STAT6;FADD;SMAD2;TGFB2;SMAD4;TGFB1;SMAD3;TGFB3;STAT1;STAT2;STAT3;NFATC1;MAPK14;NFKB1;NFKBIA;MAVS;IL6;CDK2;FAS;BAX;GRB2;KRAS;MYD88
## 5 CD86;CDKN1A;CSF2;CXCL8;TRADD;CXCL1;CXCL3;FGF2;CXCL2;ICAM1;CASP9;IKBKB;TBK1;CASP8;CCND1;MYC;AKT2;CASP3;AKT1;CCR5;JAK2;HRAS;CCR3;MAP2K1;SYK;CHUK;IFNGR1;TYK2;FOS;HLA-G;TNFRSF1A;HCK;CREB1;IRF3;IRF7;TP53;TLR3;IFNAR1;BECN1;SRC;PDGFB;PTGS2;HIF1A;RELA;C3;MAPK8;FADD;BAK1;LYN;CCR1;STAT1;STAT2;STAT3;NFATC1;MAPK14;NFKB1;MTOR;NFKBIA;RCAN1;IL6;CDK6;CDK4;FAS;BAX;CTNNB1;KRAS
## 6 SCARB1;CDKN1A;CD81;TRADD;ARAF;FASLG;IFIT1;TNF;CASP9;IKBKB;TBK1;CASP8;CCND1;MYC;AKT2;CASP3;AKT1;HRAS;MAP2K1;RSAD2;APAF1;CHUK;DDX58;TYK2;PIAS1;TNFRSF1A;CLDN4;CLDN3;OAS1;IRF3;IFNG;TRAF6;IRF7;TP53;TLR3;IFNAR1;RELA;EGFR;SOCS3;RIPK1;FADD;BAK1;LDLR;EGF;STAT1;STAT2;MX1;STAT3;NFKB1;NFKBIA;CXCL10;OCLN;MAVS;CDK6;CDK4;CDK2;FAS;BAX;CTNNB1;GRB2;KRAS## Term Overlap P.value Adjusted.P.value Old.P.value Old.Adjusted.P.value Odds.Ratio Combined.Score
## 1 cytokine activity (GO:0005125) 61/173 3.155810e-40 2.098614e-37 0 0 13.461367 1224.3673
## 2 receptor ligand activity (GO:0048018) 69/307 1.967964e-31 6.543481e-29 0 0 7.192804 508.5538
## 3 chemokine receptor binding (GO:0042379) 27/50 1.496174e-24 3.316519e-22 0 0 27.918715 1531.5964
## 4 chemokine activity (GO:0008009) 26/46 2.168043e-24 3.604371e-22 0 0 30.883871 1682.8071
## 5 cytokine receptor binding (GO:0005126) 34/105 1.501517e-21 1.997018e-19 0 0 11.459953 549.4796
## 6 kinase binding (GO:0019900) 70/461 3.129734e-21 3.468788e-19 0 0 4.411563 208.2846
## Genes
## 1 CXCL6;CSF3;CXCL9;CSF2;CXCL8;EPO;CXCL1;HMGB1;CXCL13;CXCL3;TNF;FGF2;CXCL2;CX3CL1;CXCL5;TNFSF11;TIMP1;IL10;IL11;IL15;WNT5A;ADIPOQ;IL18;NRG1;MIF;IL1A;IFNG;IL1B;CCL13;CCL11;TNFRSF11B;CCL7;CCL2;CCL1;CCL19;CCL17;CCL25;IL33;TGFB2;TGFB1;CCL22;TSLP;CCL21;TGFB3;WNT3A;CCL20;LIF;PPBP;BMP7;IL2;IL4;BMP4;CXCL10;CXCL11;BMP2;IL6;CXCL12;IL17F;INHA;CCL27;PF4
## 2 CSF3;CXCL9;CSF2;EDA;EPO;CXCL1;PRL;HMGB1;CXCL13;TNF;FGF2;CX3CL1;TNFSF13B;LGALS3;FGF7;TNFSF11;TIMP1;AMH;IL10;IL11;IL15;HGF;ADIPOQ;WNT5A;IL18;APOA1;NRG1;MIF;NGF;PGF;BTC;IL1A;IFNG;IL1B;DEFB4A;HBEGF;CCL11;PTH;PDGFB;TNFRSF11B;PTN;NTS;INS;NPY;PPY;CCL25;IL33;TGFB2;TGFB1;TSLP;TGFB3;WNT3A;BDNF;EGF;LIF;CCK;BMP7;IL2;IL4;BMP4;GH2;POMC;CXCL10;BMP2;IL6;LEP;IL17F;INHA;VIP
## 3 CX3CR1;CXCL6;CCL13;CXCL9;CXCL8;CCL11;CXCL1;CXCL13;CXCL3;CXCL2;CX3CL1;CXCL5;CCL7;CCL2;CCL1;CCL19;CCL17;CCL25;CCL22;CCL21;CCL20;PPBP;CXCL10;CXCL11;CXCL12;CCL27;PF4
## 4 CCL13;CXCL6;CXCL9;CXCL8;CCL11;CXCL1;CXCL13;CXCL3;CXCL2;CX3CL1;CXCL5;CCL7;CCL2;CCL1;CCL19;CCL17;CCL25;CCL22;CCL21;CCL20;PPBP;CXCL10;CXCL11;CXCL12;CCL27;PF4
## 5 IL21;CSF3;IL1RN;EPO;PRL;PYCARD;TNFSF11;FADD;CCL19;JAK2;IL12RB1;SMAD2;IL10;CCL25;IL11;TGFB2;SMAD3;TSLP;SYK;CCL21;LIF;IL18;MIF;TYK2;PGF;IL2;IL4;GH2;ADAM17;IL6;CXCL12;TRAF6;IL1B;TLR9
## 6 CDKN1A;TFRC;TRADD;ILK;HSPB1;PARK7;FOXM1;IKBKB;CASP9;CCND1;RAC2;CASP1;JAK2;CHIA;PARP1;RPS6;RHOG;ADAM10;IRAK3;IRAK4;RHOC;CDC25A;RHOA;RHOB;CCNA2;FGR;CEACAM1;TRAF6;VDAC1;MAPT;SQSTM1;CRK;TP53;ATF4;CEBPA;BECN1;SRC;MVP;PLG;IQGAP1;NOD2;PTN;FOXO3;HIF1A;EGFR;RELA;TTN;RELB;CDC42;CCNB1;SOCS1;IRAK1;RPS3;PTPN1;SMAD3;CAV1;PLK1;STAT3;CDC6;EEF2;IRGM;PTK2;CYLD;MAVS;CD4;TOLLIP;CTNNB1;FAS;GRB2;BCL2L1## Plot Enrichment result
p <- plotEnrich(enriched[['KEGG_2019_Human']] %>% filter(Adjusted.P.value < 0.05),
showTerms = 20, numChar = 40, y = "Count", orderBy = "P.value") +
scale_fill_gradientn(colours = rev(wes_palette("Zissou1", 10, type = "continuous")))
p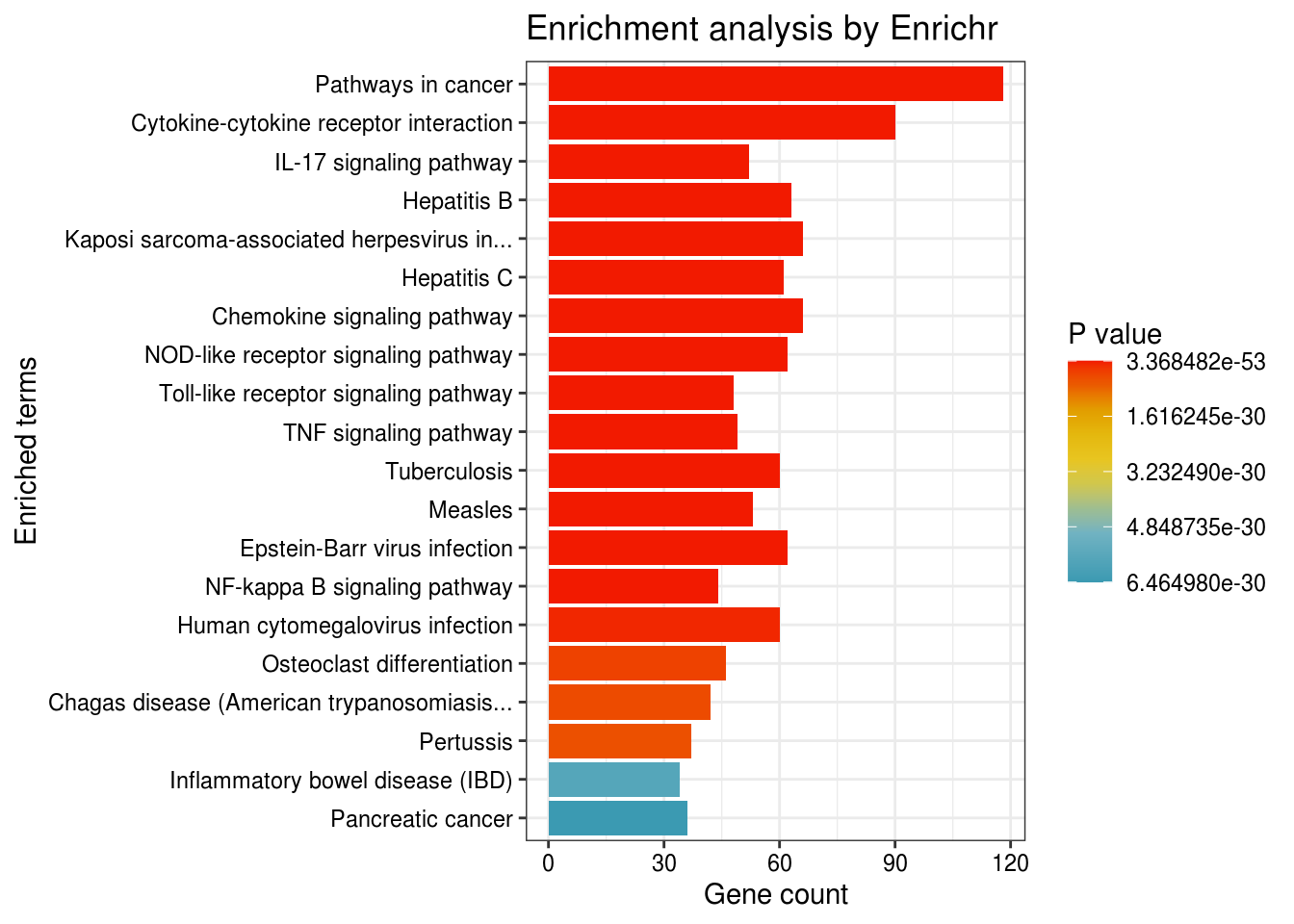
As shown above, Y axis shows the enriched terms of your input genes. X axis shows the number of input genes in the enriched terms.
5.7 Session info
## ─ Session info ──────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────
## setting value
## version R version 3.6.3 (2020-02-29)
## os Ubuntu 16.04.7 LTS
## system x86_64, linux-gnu
## ui RStudio
## language (EN)
## collate en_IN.UTF-8
## ctype en_IN.UTF-8
## tz Asia/Hong_Kong
## date 2022-11-09
## rstudio 1.1.419 (server)
## pandoc 2.7.3 @ /usr/bin/ (via rmarkdown)
##
## ─ Packages ──────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────
## ! package * version date (UTC) lib source
## abind 1.4-5 2016-07-21 [1] CRAN (R 3.6.3)
## ade4 1.7-17 2021-06-17 [1] CRAN (R 3.6.3)
## ALDEx2 * 1.18.0 2019-10-29 [1] Bioconductor
## annotate 1.64.0 2019-10-29 [1] Bioconductor
## AnnotationDbi * 1.58.0 2022-04-26 [1] Bioconductor
## ape 5.5 2021-04-25 [1] CRAN (R 3.6.3)
## assertthat 0.2.1 2019-03-21 [2] CRAN (R 3.6.3)
## backports 1.4.1 2021-12-13 [1] CRAN (R 3.6.3)
## base64enc 0.1-3 2015-07-28 [2] CRAN (R 3.6.3)
## bayesm 3.1-4 2019-10-15 [1] CRAN (R 3.6.3)
## biglm 0.9-2.1 2020-11-27 [1] CRAN (R 3.6.3)
## Biobase * 2.46.0 2019-10-29 [2] Bioconductor
## BiocGenerics * 0.32.0 2019-10-29 [2] Bioconductor
## BiocParallel * 1.20.1 2019-12-21 [2] Bioconductor
## biomformat 1.14.0 2019-10-29 [1] Bioconductor
## Biostrings 2.54.0 2019-10-29 [1] Bioconductor
## bit 4.0.4 2020-08-04 [1] CRAN (R 3.6.3)
## bit64 4.0.5 2020-08-30 [1] CRAN (R 3.6.3)
## bitops 1.0-7 2021-04-24 [1] CRAN (R 3.6.3)
## blob 1.2.2 2021-07-23 [1] CRAN (R 3.6.3)
## bookdown 0.24 2021-09-02 [1] CRAN (R 3.6.3)
## brio 1.1.3 2021-11-30 [2] CRAN (R 3.6.3)
## broom 0.7.12 2022-01-28 [1] CRAN (R 3.6.3)
## bslib 0.3.1 2021-10-06 [1] CRAN (R 3.6.3)
## cachem 1.0.5 2021-05-15 [1] CRAN (R 3.6.3)
## callr 3.7.0 2021-04-20 [2] CRAN (R 3.6.3)
## car 3.0-12 2021-11-06 [1] CRAN (R 3.6.3)
## carData 3.0-4 2020-05-22 [1] CRAN (R 3.6.3)
## caTools 1.18.2 2021-03-28 [1] CRAN (R 3.6.3)
## cellranger 1.1.0 2016-07-27 [1] CRAN (R 3.6.3)
## checkmate 2.0.0 2020-02-06 [1] CRAN (R 3.6.3)
## circlize * 0.4.13 2021-06-09 [1] CRAN (R 3.6.3)
## cli 3.1.0 2021-10-27 [1] CRAN (R 3.6.3)
## clue 0.3-59 2021-04-16 [1] CRAN (R 3.6.3)
## cluster 2.1.0 2019-06-19 [2] CRAN (R 3.6.3)
## coda 0.19-4 2020-09-30 [1] CRAN (R 3.6.3)
## codetools 0.2-16 2018-12-24 [2] CRAN (R 3.6.3)
## coin 1.4-2 2021-10-08 [1] CRAN (R 3.6.3)
## colorspace 2.0-2 2021-06-24 [1] CRAN (R 3.6.3)
## ComplexHeatmap * 2.2.0 2019-10-29 [1] Bioconductor
## compositions 2.0-2 2021-07-14 [1] CRAN (R 3.6.3)
## cowplot * 1.1.1 2020-12-30 [1] CRAN (R 3.6.3)
## crayon 1.5.0 2022-02-14 [1] CRAN (R 3.6.3)
## curl 4.3.2 2021-06-23 [2] CRAN (R 3.6.3)
## dada2 * 1.14.1 2020-02-22 [1] Bioconductor
## data.table * 1.14.0 2021-02-21 [1] CRAN (R 3.6.3)
## DBI 1.1.1 2021-01-15 [1] CRAN (R 3.6.3)
## dbplyr 2.1.1 2021-04-06 [1] CRAN (R 3.6.3)
## DelayedArray * 0.12.3 2020-04-09 [2] Bioconductor
## DelayedMatrixStats 1.8.0 2019-10-29 [1] Bioconductor
## DEoptimR 1.0-9 2021-05-24 [1] CRAN (R 3.6.3)
## desc 1.4.1 2022-03-06 [2] CRAN (R 3.6.3)
## DESeq2 * 1.26.0 2019-10-29 [1] Bioconductor
## devtools 2.4.3 2021-11-30 [1] CRAN (R 3.6.3)
## digest 0.6.29 2021-12-01 [1] CRAN (R 3.6.3)
## dplyr * 1.0.6 2021-05-05 [1] CRAN (R 3.6.3)
## edgeR 3.28.1 2020-02-26 [1] Bioconductor
## ellipsis 0.3.2 2021-04-29 [1] CRAN (R 3.6.3)
## EnhancedVolcano * 1.4.0 2019-10-29 [1] Bioconductor
## enrichR * 3.0 2021-02-02 [1] CRAN (R 3.6.3)
## evaluate 0.15 2022-02-18 [2] CRAN (R 3.6.3)
## fansi 1.0.2 2022-01-14 [1] CRAN (R 3.6.3)
## farver 2.1.0 2021-02-28 [2] CRAN (R 3.6.3)
## fastmap 1.1.0 2021-01-25 [1] CRAN (R 3.6.3)
## fdrtool 1.2.17 2021-11-13 [1] CRAN (R 3.6.3)
## forcats * 0.5.1 2021-01-27 [1] CRAN (R 3.6.3)
## foreach 1.5.2 2022-02-02 [2] CRAN (R 3.6.3)
## foreign 0.8-75 2020-01-20 [2] CRAN (R 3.6.3)
## formatR 1.12 2022-03-31 [2] CRAN (R 3.6.3)
## Formula 1.2-4 2020-10-16 [1] CRAN (R 3.6.3)
## fs 1.5.2 2021-12-08 [1] CRAN (R 3.6.3)
## futile.logger 1.4.3 2016-07-10 [2] CRAN (R 3.6.3)
## futile.options 1.0.1 2018-04-20 [2] CRAN (R 3.6.3)
## genefilter 1.68.0 2019-10-29 [1] Bioconductor
## geneplotter 1.64.0 2019-10-29 [1] Bioconductor
## generics 0.1.2 2022-01-31 [1] CRAN (R 3.6.3)
## GenomeInfoDb * 1.22.1 2020-03-27 [2] Bioconductor
## GenomeInfoDbData 1.2.2 2020-08-24 [2] Bioconductor
## GenomicAlignments 1.22.1 2019-11-12 [1] Bioconductor
## GenomicRanges * 1.38.0 2019-10-29 [2] Bioconductor
## getopt 1.20.3 2019-03-22 [1] CRAN (R 3.6.3)
## GetoptLong 1.0.5 2020-12-15 [1] CRAN (R 3.6.3)
## GGally * 2.1.2 2021-06-21 [1] CRAN (R 3.6.3)
## ggbipart * 0.1.2 2022-07-20 [1] Github (pedroj/bipartite_plots@162f577)
## ggExtra * 0.9 2019-08-27 [1] CRAN (R 3.6.3)
## ggplot2 * 3.3.5 2021-06-25 [1] CRAN (R 3.6.3)
## ggpubr * 0.4.0 2020-06-27 [1] CRAN (R 3.6.3)
## ggrepel * 0.9.1 2021-01-15 [2] CRAN (R 3.6.3)
## ggsci * 2.9 2018-05-14 [1] CRAN (R 3.6.3)
## ggsignif 0.6.3 2021-09-09 [1] CRAN (R 3.6.3)
## glmnet 4.1-2 2021-06-24 [1] CRAN (R 3.6.3)
## GlobalOptions 0.1.2 2020-06-10 [1] CRAN (R 3.6.3)
## glue 1.6.1 2022-01-22 [1] CRAN (R 3.6.3)
## GMPR 0.1.3 2021-05-17 [1] local
## gplots 3.1.1 2020-11-28 [1] CRAN (R 3.6.3)
## graph 1.64.0 2019-10-29 [1] Bioconductor
## gridExtra 2.3 2017-09-09 [2] CRAN (R 3.6.3)
## gtable 0.3.0 2019-03-25 [2] CRAN (R 3.6.3)
## gtools 3.9.2 2021-06-06 [1] CRAN (R 3.6.3)
## haven 2.4.1 2021-04-23 [1] CRAN (R 3.6.3)
## highr 0.9 2021-04-16 [1] CRAN (R 3.6.3)
## Hmisc 4.5-0 2021-02-28 [1] CRAN (R 3.6.3)
## hms 1.1.1 2021-09-26 [1] CRAN (R 3.6.3)
## htmlTable 2.3.0 2021-10-12 [1] CRAN (R 3.6.3)
## htmltools 0.5.2 2021-08-25 [1] CRAN (R 3.6.3)
## htmlwidgets 1.5.4 2021-09-08 [2] CRAN (R 3.6.3)
## httpuv 1.6.1 2021-05-07 [1] CRAN (R 3.6.3)
## httr 1.4.3 2022-05-04 [2] CRAN (R 3.6.3)
## hwriter 1.3.2 2014-09-10 [1] CRAN (R 3.6.3)
## igraph 1.3.1 2022-04-20 [2] CRAN (R 3.6.3)
## IHW 1.14.0 2019-10-29 [1] Bioconductor
## IRanges * 2.20.2 2020-01-13 [2] Bioconductor
## iterators 1.0.14 2022-02-05 [2] CRAN (R 3.6.3)
## jpeg 0.1-9 2021-07-24 [1] CRAN (R 3.6.3)
## jquerylib 0.1.4 2021-04-26 [1] CRAN (R 3.6.3)
## jsonlite 1.8.0 2022-02-22 [2] CRAN (R 3.6.3)
## KEGGgraph 1.46.0 2019-10-29 [1] Bioconductor
## KEGGREST 1.26.1 2019-11-06 [1] Bioconductor
## KernSmooth 2.23-16 2019-10-15 [2] CRAN (R 3.6.3)
## knitr 1.36 2021-09-29 [1] CRAN (R 3.6.3)
## labeling 0.4.2 2020-10-20 [2] CRAN (R 3.6.3)
## lambda.r 1.2.4 2019-09-18 [2] CRAN (R 3.6.3)
## later 1.3.0 2021-08-18 [2] CRAN (R 3.6.3)
## lattice * 0.20-38 2018-11-04 [2] CRAN (R 3.6.3)
## latticeExtra 0.6-29 2019-12-19 [1] CRAN (R 3.6.3)
## lazyeval 0.2.2 2019-03-15 [2] CRAN (R 3.6.3)
## libcoin 1.0-9 2021-09-27 [1] CRAN (R 3.6.3)
## lifecycle 1.0.1 2021-09-24 [1] CRAN (R 3.6.3)
## limma 3.42.2 2020-02-03 [2] Bioconductor
## locfit 1.5-9.4 2020-03-25 [1] CRAN (R 3.6.3)
## lpsymphony 1.14.0 2019-10-29 [1] Bioconductor (R 3.6.3)
## lubridate 1.7.10 2021-02-26 [1] CRAN (R 3.6.3)
## Maaslin2 1.7.3 2022-03-23 [1] Github (biobakery/maaslin2@8d090e4)
## magrittr * 2.0.2 2022-01-26 [1] CRAN (R 3.6.3)
## MASS 7.3-54 2021-05-03 [1] CRAN (R 3.6.3)
## Matrix 1.3-4 2021-06-01 [1] CRAN (R 3.6.3)
## matrixStats * 0.60.0 2021-07-26 [1] CRAN (R 3.6.3)
## mbzinb 0.2 2021-06-23 [1] local
## memoise 2.0.1 2021-11-26 [2] CRAN (R 3.6.3)
## metagenomeSeq 1.28.2 2020-02-03 [1] Bioconductor
## metamicrobiomeR 1.1 2021-02-03 [1] local
## mgcv 1.8-31 2019-11-09 [2] CRAN (R 3.6.3)
## microbiome 1.8.0 2019-10-29 [1] Bioconductor
## mime 0.12 2021-09-28 [2] CRAN (R 3.6.3)
## miniUI 0.1.1.1 2018-05-18 [1] CRAN (R 3.6.3)
## modelr 0.1.8 2020-05-19 [1] CRAN (R 3.6.3)
## modeltools 0.2-23 2020-03-05 [1] CRAN (R 3.6.3)
## multcomp 1.4-17 2021-04-29 [1] CRAN (R 3.6.3)
## multtest 2.42.0 2019-10-29 [2] Bioconductor
## munsell 0.5.0 2018-06-12 [2] CRAN (R 3.6.3)
## mvtnorm 1.1-3 2021-10-08 [1] CRAN (R 3.6.3)
## network * 1.17.1 2021-06-14 [1] CRAN (R 3.6.3)
## nlme 3.1-144 2020-02-06 [2] CRAN (R 3.6.3)
## nnet 7.3-12 2016-02-02 [2] CRAN (R 3.6.3)
## optparse 1.7.1 2021-10-08 [1] CRAN (R 3.6.3)
## org.Hs.eg.db * 3.10.0 2021-12-08 [1] Bioconductor
## pathview * 1.26.0 2019-10-29 [1] Bioconductor
## pcaPP 1.9-74 2021-04-23 [1] CRAN (R 3.6.3)
## permute * 0.9-5 2019-03-12 [1] CRAN (R 3.6.3)
## phyloseq * 1.30.0 2019-10-29 [1] Bioconductor
## pillar 1.7.0 2022-02-01 [1] CRAN (R 3.6.3)
## pkgbuild 1.3.1 2021-12-20 [2] CRAN (R 3.6.3)
## pkgconfig 2.0.3 2019-09-22 [2] CRAN (R 3.6.3)
## pkgload 1.2.4 2021-11-30 [2] CRAN (R 3.6.3)
## plotly * 4.10.0 2021-10-09 [1] CRAN (R 3.6.3)
## plyr 1.8.7 2022-03-24 [2] CRAN (R 3.6.3)
## png 0.1-7 2013-12-03 [1] CRAN (R 3.6.3)
## prettyunits 1.1.1 2020-01-24 [2] CRAN (R 3.6.3)
## processx 3.5.3 2022-03-25 [2] CRAN (R 3.6.3)
## promises 1.2.0.1 2021-02-11 [2] CRAN (R 3.6.3)
## protoclust 1.6.3 2019-01-31 [1] CRAN (R 3.6.3)
## ps 1.7.0 2022-04-23 [2] CRAN (R 3.6.3)
## pscl 1.5.5 2020-03-07 [1] CRAN (R 3.6.3)
## purrr * 0.3.4 2020-04-17 [2] CRAN (R 3.6.3)
## qvalue 2.18.0 2019-10-29 [1] Bioconductor
## R6 2.5.1 2021-08-19 [1] CRAN (R 3.6.3)
## RAIDA 1.0 2021-06-23 [1] local
## ranacapa 0.1.0 2021-06-18 [1] Github (gauravsk/ranacapa@58c0cab)
## RColorBrewer * 1.1-3 2022-04-03 [2] CRAN (R 3.6.3)
## Rcpp * 1.0.7 2021-07-07 [1] CRAN (R 3.6.3)
## RcppParallel 5.1.4 2021-05-04 [1] CRAN (R 3.6.3)
## RCurl 1.98-1.6 2022-02-08 [2] CRAN (R 3.6.3)
## readr * 2.0.0 2021-07-20 [1] CRAN (R 3.6.3)
## readxl * 1.3.1 2019-03-13 [1] CRAN (R 3.6.3)
## remotes 2.4.2 2021-11-30 [1] CRAN (R 3.6.3)
## reprex 2.0.1 2021-08-05 [1] CRAN (R 3.6.3)
## reshape 0.8.9 2022-04-12 [1] CRAN (R 3.6.3)
## reshape2 * 1.4.4 2020-04-09 [2] CRAN (R 3.6.3)
## Rgraphviz 2.30.0 2019-10-29 [1] Bioconductor
## rhdf5 2.30.1 2019-11-26 [1] Bioconductor
## Rhdf5lib 1.8.0 2019-10-29 [1] Bioconductor
## rJava 1.0-5 2021-09-24 [1] CRAN (R 3.6.3)
## rjson 0.2.20 2018-06-08 [1] CRAN (R 3.6.3)
## R rlang 1.0.2 <NA> [2] <NA>
## rmarkdown 2.11 2021-09-14 [1] CRAN (R 3.6.3)
## robustbase 0.93-9 2021-09-27 [1] CRAN (R 3.6.3)
## rpart 4.1-15 2019-04-12 [2] CRAN (R 3.6.3)
## rprojroot 2.0.2 2020-11-15 [1] CRAN (R 3.6.3)
## Rsamtools 2.2.3 2020-02-23 [1] Bioconductor
## RSQLite 2.2.7 2021-04-22 [1] CRAN (R 3.6.3)
## rstatix 0.7.0 2021-02-13 [1] CRAN (R 3.6.3)
## rstudioapi 0.13 2020-11-12 [2] CRAN (R 3.6.3)
## Rtsne 0.15 2018-11-10 [1] CRAN (R 3.6.3)
## rvest 1.0.2 2021-10-16 [1] CRAN (R 3.6.3)
## S4Vectors * 0.24.4 2020-04-09 [2] Bioconductor
## sandwich 3.0-1 2021-05-18 [1] CRAN (R 3.6.3)
## sass 0.4.0 2021-05-12 [1] CRAN (R 3.6.3)
## scales 1.2.0 2022-04-13 [2] CRAN (R 3.6.3)
## seqinr * 4.2-8 2021-06-09 [1] CRAN (R 3.6.3)
## sessioninfo 1.2.2 2021-12-06 [2] CRAN (R 3.6.3)
## shape 1.4.6 2021-05-19 [1] CRAN (R 3.6.3)
## shiny 1.7.1 2021-10-02 [1] CRAN (R 3.6.3)
## ShortRead 1.44.3 2020-02-03 [1] Bioconductor
## slam 0.1-49 2021-11-17 [1] CRAN (R 3.6.3)
## sna 2.6 2020-10-06 [1] CRAN (R 3.6.3)
## statnet.common 4.5.0 2021-06-05 [1] CRAN (R 3.6.3)
## stringi 1.7.4 2021-08-25 [1] CRAN (R 3.6.3)
## stringr * 1.4.0 2019-02-10 [2] CRAN (R 3.6.3)
## SummarizedExperiment * 1.16.1 2019-12-19 [2] Bioconductor
## survival 3.1-8 2019-12-03 [2] CRAN (R 3.6.3)
## tensorA 0.36.2 2020-11-19 [1] CRAN (R 3.6.3)
## testthat 3.1.4 2022-04-26 [2] CRAN (R 3.6.3)
## textshape 1.7.3 2021-05-28 [1] CRAN (R 3.6.3)
## TH.data 1.1-0 2021-09-27 [1] CRAN (R 3.6.3)
## tibble * 3.1.6 2021-11-07 [1] CRAN (R 3.6.3)
## tidyr * 1.2.0 2022-02-01 [1] CRAN (R 3.6.3)
## tidyselect 1.1.1 2021-04-30 [1] CRAN (R 3.6.3)
## tidyverse * 1.3.1 2021-04-15 [1] CRAN (R 3.6.3)
## tzdb 0.2.0 2021-10-27 [1] CRAN (R 3.6.3)
## UpSetR 1.4.0 2019-05-22 [1] CRAN (R 3.6.3)
## usethis 2.1.6 2022-05-25 [2] CRAN (R 3.6.3)
## utf8 1.2.2 2021-07-24 [1] CRAN (R 3.6.3)
## vctrs 0.3.8 2021-04-29 [1] CRAN (R 3.6.3)
## vegan * 2.5-7 2020-11-28 [1] CRAN (R 3.6.3)
## VennDiagram 1.7.1 2021-12-02 [1] CRAN (R 3.6.3)
## viridisLite 0.4.0 2021-04-13 [2] CRAN (R 3.6.3)
## vroom 1.5.7 2021-11-30 [1] CRAN (R 3.6.3)
## wesanderson * 0.3.6.9000 2021-07-21 [1] Github (karthik/wesanderson@651c944)
## withr 2.4.3 2021-11-30 [1] CRAN (R 3.6.3)
## Wrench 1.4.0 2019-10-29 [1] Bioconductor
## xfun 0.23 2021-05-15 [1] CRAN (R 3.6.3)
## xlsx * 0.6.5 2020-11-10 [1] CRAN (R 3.6.3)
## xlsxjars 0.6.1 2014-08-22 [1] CRAN (R 3.6.3)
## XMAS * 0.0.0.9000 2022-03-23 [1] local
## XMAS2 2.1.8.3 2022-11-08 [2] local
## XML 3.99-0.3 2020-01-20 [1] CRAN (R 3.6.3)
## xml2 1.3.3 2021-11-30 [2] CRAN (R 3.6.3)
## xtable 1.8-4 2019-04-21 [1] CRAN (R 3.6.3)
## XVector 0.26.0 2019-10-29 [2] Bioconductor
## xviz * 1.1.0 2021-01-14 [1] local
## yaml 2.2.2 2022-01-25 [1] CRAN (R 3.6.3)
## zlibbioc 1.32.0 2019-10-29 [2] Bioconductor
## zoo 1.8-9 2021-03-09 [1] CRAN (R 3.6.3)
##
## [1] /share/home/tongbangzhuo/R/x86_64-pc-linux-gnu-library/3.6
## [2] /opt/R-3.6.3/lib/R/library
##
## R ── Package was removed from disk.
##
## ─────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────────