Chapter 3 HAllA
For any questions about this chapter, please contact zhaoyifan (zhaoyifan@xbiome.com)
3.1 Introduction
HAllA (Hierarchical All-against-All association) 是一个针对多维度、异质型数据集的多重关联分析工具。可用于连续型/分类型数值数据,且在同质型(homogeneous)数据集(所有数值皆相同类型,例如:RNA-seq 基因表現)与异质型(heterogeneous)数据集(不同单位或类型的数值,例如:病患临床指标)皆可高效分析。在目前微生物研究中,HAllA是一個常用于微生物组-代谢、微生物组-临床指标、微生物组-转录组(基因表現、miRNA、GO) 等关联分析的重要工具。

HAllA workflow
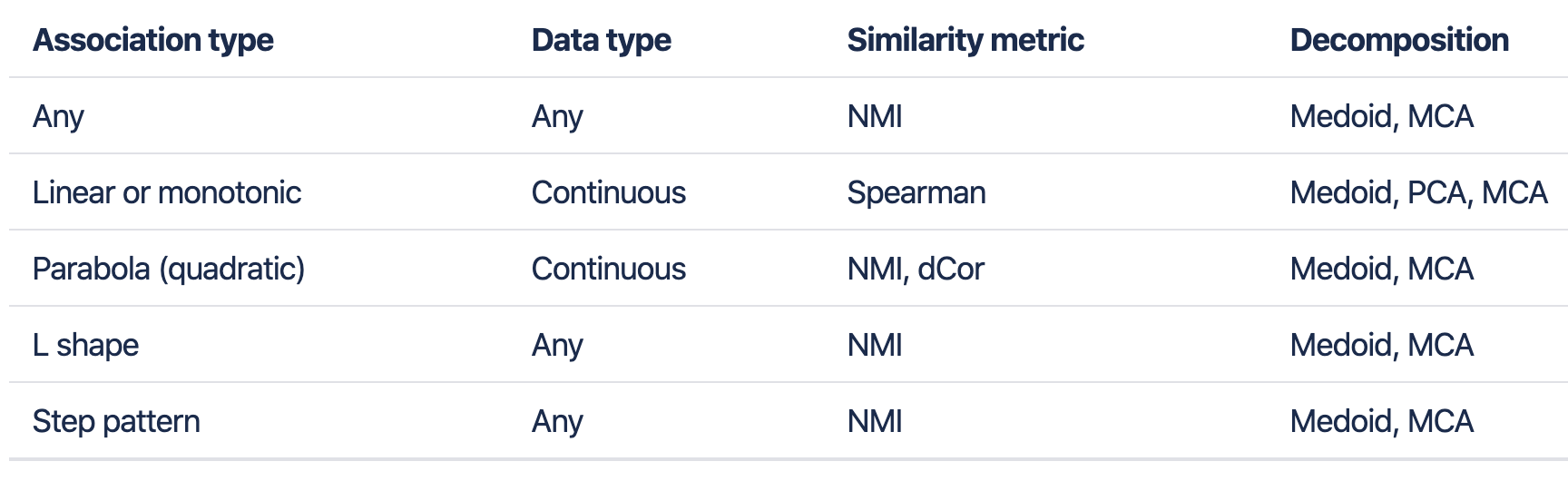
3.1.1 Available pairwise distance metrics
spearman (default for continuous data)
pearson
mi (A set of properties of Mutual Information result from definition. default for mixed/categorical data)
nmi (Normalized Mutual Information)
xicor (Chatterjee correlation, uses rank differences to assess the degree to which one variable is a measurable function of another,适用于离散、非线性关系数据)
dcor (measures not necessarily linear dependency of two random variables with possibly different dimensions)
HAllA correlation methods
3.1.2 Running procedures
3.1.2.1 step1
The pairwise similarity matrix between all features in X and Y is computed with a specified similarity measure, such as Spearman correlation and normalized mutual information (NMI). This step then generates the p-value and q-value tables.
Note that for handling heterogeneous data, all continuous features are first discretized into bins using a specified binning method.
3.1.2.2 step2
Hierarchical clustering on the features in each dataset is performed using the converted similarity measure used in step 1. It produces a tree for each dataset.
3.1.2.3 step3
Finding densely-associated blocks (pseudocode):
def find_densely_associated_blocks(x, y):
x_features = all features in x
y_features = all features in y
if is_densely_associated(x_features, y_features):
report block and terminate
else:
# bifurcate one according to Gini impurities of the splits
x_branches, y_branches = bifurcate_one_of(x, y)
if both x and y are leaves:
terminate
for each x_branch and y_branch in x_branches and y_branches:
find_densely_associated_blocks(x_branch, y_branch)
initial function call: find_densely_associated_blocks(X_root, Y_root)For example, given two datasets of X (features: X1, X2, X3, X4, X5) and Y (features: Y1, Y2, Y3, Y4) both hierarchically clustered in X tree and Y tree, the algorithm first evaluates the roots of both trees and checks if the block consisting of all features of X and Y are densely-associated (if %significance (%reject) >= (1 - FNR)%).
If the block is not densely-associated, the algorithm would bifurcate one of the trees. It would pick one of:
·[X1 X2][X3 X4 X5] >< [Y1 Y2 Y3 Y4] or
·[X1 X2 X3 X4 X5] >< [Y1 Y2 Y3][Y4]
based on the Gini impurity of the splits (pick the split that produces a lower weighted Gini impurity),
Once it picks the split with the lower impurity (let’s say the first split), it will iteratively evaluate the branches:
·find densely-associated blocks in [X1 X2] vs [Y1 Y2 Y3 Y4], and
·find densely-associated blocks in [X3 X4 X5] vs [Y1 Y2 Y3 Y4]
and keep going until it terminates.
3.2 Run HALLA
Run HALLA on remote server
3.3 Result of HALLA
HAllA result after WGCNA
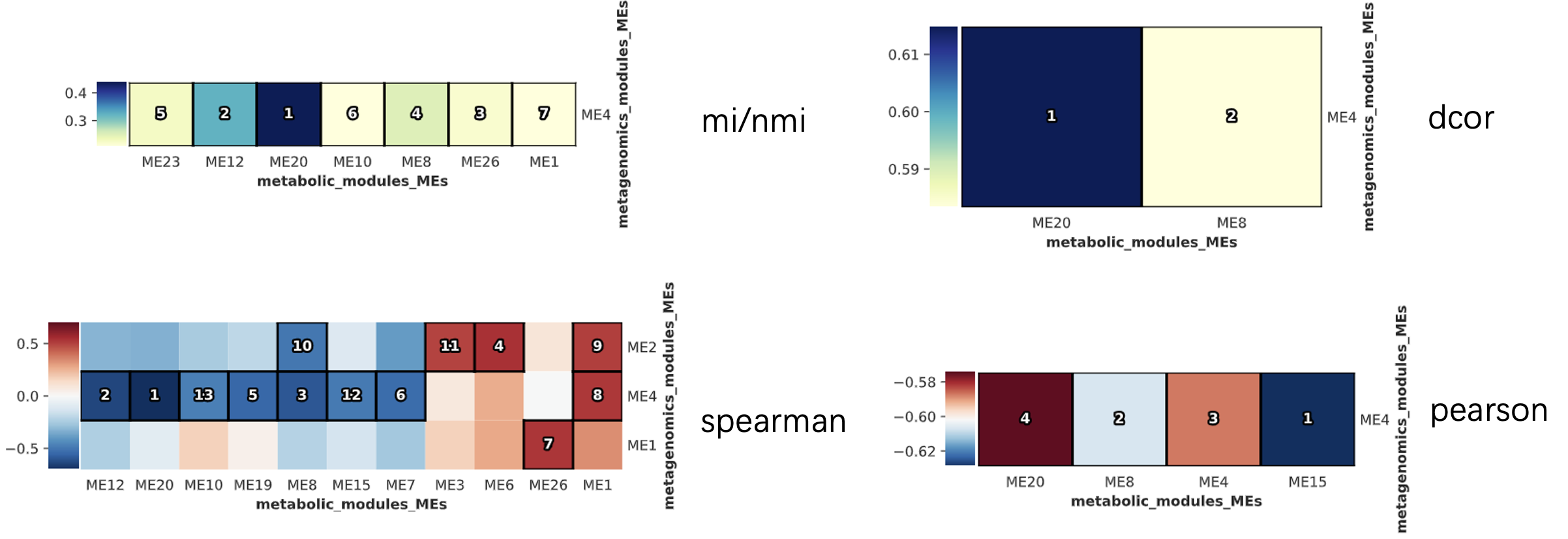
Correlation with modules